# pyMSAviz



[](https://pypi.python.org/pypi/pymsaviz)
[](https://anaconda.org/bioconda/pymsaviz)
[](https://github.com/moshi4/pyMSAviz/actions/workflows/ci.yml)
## Table of contents
- [Overview](#overview)
- [Installation](#installation)
- [API Usage](#api-usage)
- [CLI Usage](#cli-usage)
## Overview
pyMSAviz is a MSA(Multiple Sequence Alignment) visualization python package for sequence analysis implemented based on matplotlib.
This package is developed for the purpose of easily and beautifully plotting MSA in Python.
It also implements the functionality to add markers, text annotations, highlights to specific positions and ranges in MSA.
pyMSAviz was developed inspired by [Jalview](https://www.jalview.org/) and [ggmsa](https://github.com/YuLab-SMU/ggmsa).
More detailed documentation is available [here](https://moshi4.github.io/pyMSAviz/).
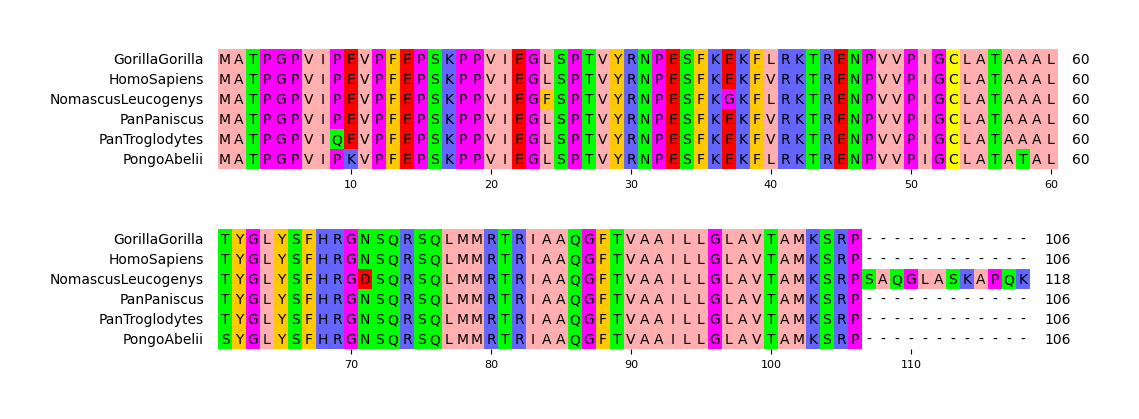
**Fig.1 Simple visualization result**
**Fig.2 Customized visualization result**
## Installation
`Python 3.9 or later` is required for installation.
**Install PyPI package:**
pip install pymsaviz
**Install bioconda package:**
conda install -c conda-forge -c bioconda pymsaviz
## API Usage
Only simple example usage is described in this section.
For more details, please see [Getting Started](https://moshi4.github.io/pyMSAviz/getting_started/) and [API Docs](https://moshi4.github.io/pyMSAviz/api-docs/msaviz/).
### API Example
#### API Example 1
```python
from pymsaviz import MsaViz, get_msa_testdata
msa_file = get_msa_testdata("HIGD2A.fa")
mv = MsaViz(msa_file, wrap_length=60, show_count=True)
mv.savefig("api_example01.png")
```

#### API Example 2
```python
from pymsaviz import MsaViz, get_msa_testdata
msa_file = get_msa_testdata("MRGPRG.fa")
mv = MsaViz(msa_file, color_scheme="Taylor", wrap_length=80, show_grid=True, show_consensus=True)
mv.savefig("api_example02.png")
```
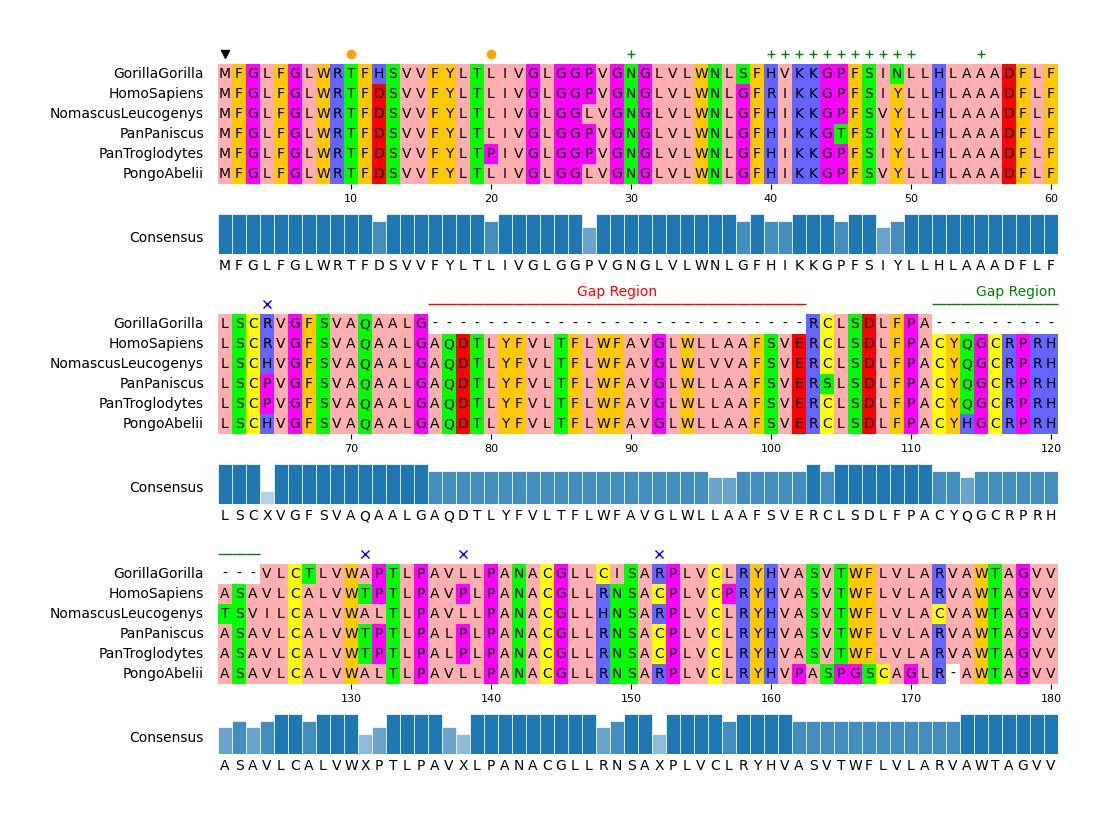
#### API Example 3
```python
from pymsaviz import MsaViz, get_msa_testdata
msa_file = get_msa_testdata("MRGPRG.fa")
mv = MsaViz(msa_file, end=180, wrap_length=60, show_consensus=True)
# Extract MSA positions less than 50% consensus identity
pos_ident_less_than_50 = []
ident_list = mv._get_consensus_identity_list()
for pos, ident in enumerate(ident_list, 1):
if ident <= 50:
pos_ident_less_than_50.append(pos)
# Add markers
mv.add_markers([1])
mv.add_markers([10, 20], color="orange", marker="o")
mv.add_markers([30, (40, 50), 55], color="green", marker="+")
mv.add_markers(pos_ident_less_than_50, marker="x", color="blue")
# Add text annotations
mv.add_text_annotation((76, 102), "Gap Region", text_color="red", range_color="red")
mv.add_text_annotation((112, 123), "Gap Region", text_color="green", range_color="green")
mv.savefig("api_example03.png")
```

## CLI Usage
pyMSAviz provides simple MSA visualization CLI.
### Basic Command
pymsaviz -i [MSA file] -o [MSA visualization file]
### Options
$ pymsaviz --help
usage: pymsaviz [options] -i msa.fa -o msa_viz.png
MSA(Multiple Sequence Alignment) visualization CLI tool
optional arguments:
-i I, --infile I Input MSA file
-o O, --outfile O Output MSA visualization file (*.png|*.jpg|*.svg|*.pdf)
--format MSA file format (Default: 'fasta')
--color_scheme Color scheme (Default: 'Zappo')
--start Start position of MSA visualization (Default: 1)
--end End position of MSA visualization (Default: 'MSA Length')
--wrap_length Wrap length (Default: None)
--wrap_space_size Space size between wrap MSA plot area (Default: 3.0)
--label_type Label type ('id'[default]|'description')
--show_grid Show grid (Default: OFF)
--show_count Show seq char count without gap on right side (Default: OFF)
--show_consensus Show consensus sequence (Default: OFF)
--consensus_color Consensus identity bar color (Default: '#1f77b4')
--consensus_size Consensus identity bar height size (Default: 2.0)
--sort Sort MSA order by NJ tree constructed from MSA distance matrix (Default: OFF)
--dpi Figure DPI (Default: 300)
-v, --version Print version information
-h, --help Show this help message and exit
Available Color Schemes:
['Clustal', 'Zappo', 'Taylor', 'Flower', 'Blossom', 'Sunset', 'Ocean', 'Hydrophobicity', 'HelixPropensity', 'StrandPropensity', 'TurnPropensity', 'BuriedIndex', 'Nucleotide', 'Purine/Pyrimidine', 'Identity', 'None']
### CLI Example
Click [here](https://github.com/moshi4/pyMSAviz/raw/main/example/example.zip) to download example MSA files.
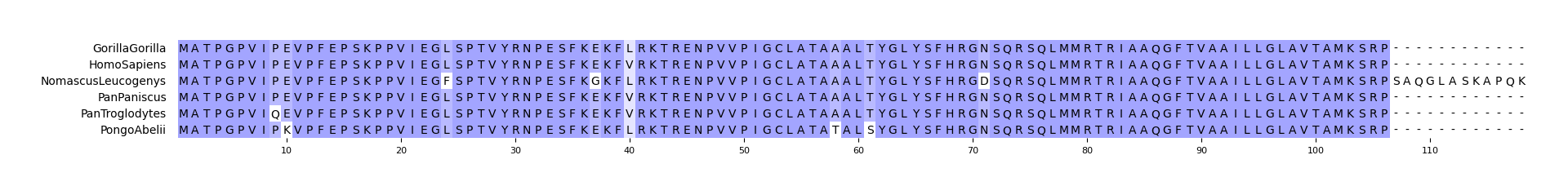
#### CLI Example 1
pymsaviz -i ./example/HIGD2A.fa -o cli_example01.png --color_scheme Identity
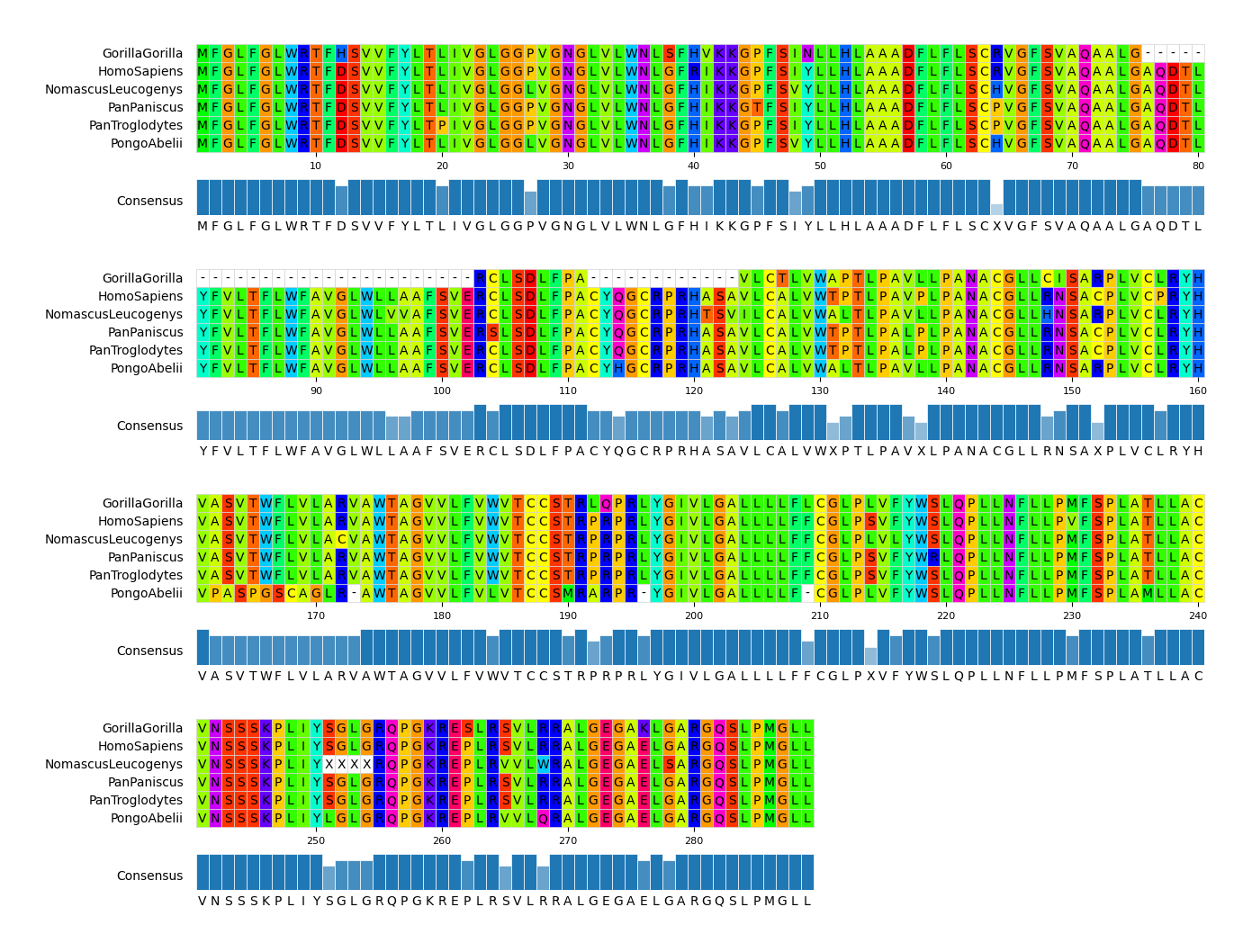
#### CLI Example 2
pymsaviz -i ./example/MRGPRG.fa -o cli_example02.png --wrap_length 80 \
--color_scheme Taylor --show_consensus --show_count
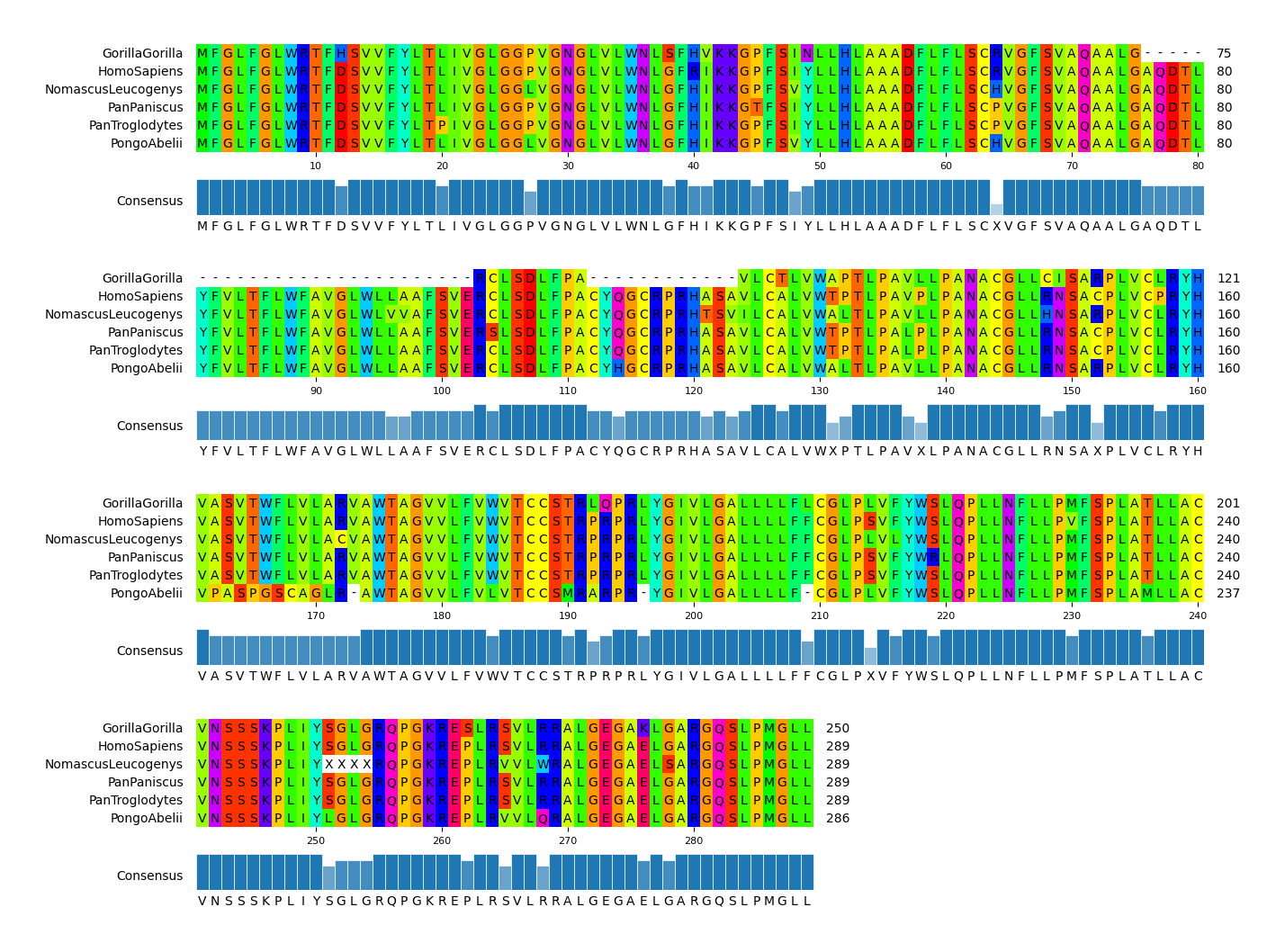
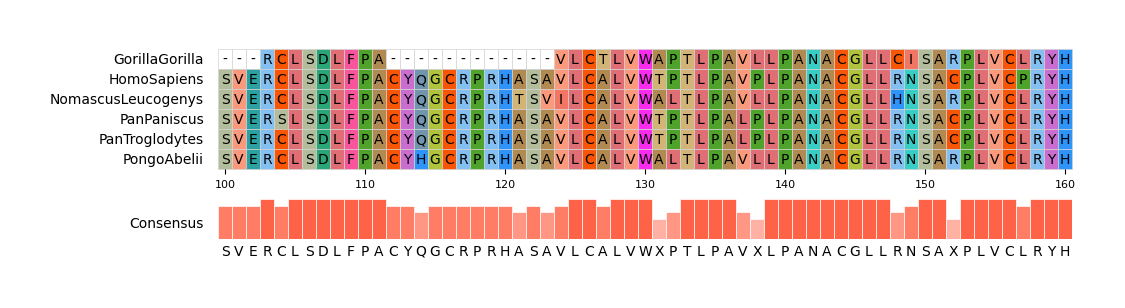
#### CLI Example 3
pymsaviz -i ./example/MRGPRG.fa -o cli_example03.png --start 100 --end 160 \
--color_scheme Flower --show_grid --show_consensus --consensus_color tomato
Raw data
{
"_id": null,
"home_page": null,
"name": "pyMSAviz",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.9",
"maintainer_email": null,
"keywords": "bioinformatics, matplotlib, sequence-alignment, sequence-analysis, visualization",
"author": "moshi4",
"author_email": null,
"download_url": "https://files.pythonhosted.org/packages/f0/55/a7ffe31c8d389f5eed61713b226abb0a5f57b55ffd4e4c686b581b54f28c/pymsaviz-0.5.0.tar.gz",
"platform": null,
"description": "# pyMSAviz\n\n\n\n\n[](https://pypi.python.org/pypi/pymsaviz)\n[](https://anaconda.org/bioconda/pymsaviz)\n[](https://github.com/moshi4/pyMSAviz/actions/workflows/ci.yml)\n\n## Table of contents\n\n- [Overview](#overview)\n- [Installation](#installation)\n- [API Usage](#api-usage)\n- [CLI Usage](#cli-usage)\n\n## Overview\n\npyMSAviz is a MSA(Multiple Sequence Alignment) visualization python package for sequence analysis implemented based on matplotlib.\nThis package is developed for the purpose of easily and beautifully plotting MSA in Python.\nIt also implements the functionality to add markers, text annotations, highlights to specific positions and ranges in MSA.\npyMSAviz was developed inspired by [Jalview](https://www.jalview.org/) and [ggmsa](https://github.com/YuLab-SMU/ggmsa).\nMore detailed documentation is available [here](https://moshi4.github.io/pyMSAviz/).\n\n \n**Fig.1 Simple visualization result**\n\n \n**Fig.2 Customized visualization result**\n\n## Installation\n\n`Python 3.9 or later` is required for installation.\n\n**Install PyPI package:**\n\n pip install pymsaviz\n\n**Install bioconda package:**\n\n conda install -c conda-forge -c bioconda pymsaviz\n\n## API Usage\n\nOnly simple example usage is described in this section.\nFor more details, please see [Getting Started](https://moshi4.github.io/pyMSAviz/getting_started/) and [API Docs](https://moshi4.github.io/pyMSAviz/api-docs/msaviz/).\n\n### API Example\n\n#### API Example 1\n\n```python\nfrom pymsaviz import MsaViz, get_msa_testdata\n\nmsa_file = get_msa_testdata(\"HIGD2A.fa\")\nmv = MsaViz(msa_file, wrap_length=60, show_count=True)\nmv.savefig(\"api_example01.png\")\n```\n\n \n\n#### API Example 2\n\n```python\nfrom pymsaviz import MsaViz, get_msa_testdata\n\nmsa_file = get_msa_testdata(\"MRGPRG.fa\")\nmv = MsaViz(msa_file, color_scheme=\"Taylor\", wrap_length=80, show_grid=True, show_consensus=True)\nmv.savefig(\"api_example02.png\")\n```\n\n \n\n#### API Example 3\n\n```python\nfrom pymsaviz import MsaViz, get_msa_testdata\n\nmsa_file = get_msa_testdata(\"MRGPRG.fa\")\nmv = MsaViz(msa_file, end=180, wrap_length=60, show_consensus=True)\n\n# Extract MSA positions less than 50% consensus identity\npos_ident_less_than_50 = []\nident_list = mv._get_consensus_identity_list()\nfor pos, ident in enumerate(ident_list, 1):\n if ident <= 50:\n pos_ident_less_than_50.append(pos)\n\n# Add markers\nmv.add_markers([1])\nmv.add_markers([10, 20], color=\"orange\", marker=\"o\")\nmv.add_markers([30, (40, 50), 55], color=\"green\", marker=\"+\")\nmv.add_markers(pos_ident_less_than_50, marker=\"x\", color=\"blue\")\n# Add text annotations\nmv.add_text_annotation((76, 102), \"Gap Region\", text_color=\"red\", range_color=\"red\")\nmv.add_text_annotation((112, 123), \"Gap Region\", text_color=\"green\", range_color=\"green\")\n\nmv.savefig(\"api_example03.png\")\n```\n\n \n\n## CLI Usage\n\npyMSAviz provides simple MSA visualization CLI.\n\n### Basic Command\n\n pymsaviz -i [MSA file] -o [MSA visualization file]\n\n### Options\n\n $ pymsaviz --help\n usage: pymsaviz [options] -i msa.fa -o msa_viz.png\n\n MSA(Multiple Sequence Alignment) visualization CLI tool\n\n optional arguments:\n -i I, --infile I Input MSA file\n -o O, --outfile O Output MSA visualization file (*.png|*.jpg|*.svg|*.pdf)\n --format MSA file format (Default: 'fasta')\n --color_scheme Color scheme (Default: 'Zappo')\n --start Start position of MSA visualization (Default: 1)\n --end End position of MSA visualization (Default: 'MSA Length')\n --wrap_length Wrap length (Default: None)\n --wrap_space_size Space size between wrap MSA plot area (Default: 3.0)\n --label_type Label type ('id'[default]|'description')\n --show_grid Show grid (Default: OFF)\n --show_count Show seq char count without gap on right side (Default: OFF)\n --show_consensus Show consensus sequence (Default: OFF)\n --consensus_color Consensus identity bar color (Default: '#1f77b4')\n --consensus_size Consensus identity bar height size (Default: 2.0)\n --sort Sort MSA order by NJ tree constructed from MSA distance matrix (Default: OFF)\n --dpi Figure DPI (Default: 300)\n -v, --version Print version information\n -h, --help Show this help message and exit\n\n Available Color Schemes:\n ['Clustal', 'Zappo', 'Taylor', 'Flower', 'Blossom', 'Sunset', 'Ocean', 'Hydrophobicity', 'HelixPropensity', 'StrandPropensity', 'TurnPropensity', 'BuriedIndex', 'Nucleotide', 'Purine/Pyrimidine', 'Identity', 'None']\n\n### CLI Example\n\nClick [here](https://github.com/moshi4/pyMSAviz/raw/main/example/example.zip) to download example MSA files. \n\n#### CLI Example 1\n\n pymsaviz -i ./example/HIGD2A.fa -o cli_example01.png --color_scheme Identity\n\n \n\n#### CLI Example 2\n\n pymsaviz -i ./example/MRGPRG.fa -o cli_example02.png --wrap_length 80 \\\n --color_scheme Taylor --show_consensus --show_count\n\n \n\n#### CLI Example 3\n\n pymsaviz -i ./example/MRGPRG.fa -o cli_example03.png --start 100 --end 160 \\\n --color_scheme Flower --show_grid --show_consensus --consensus_color tomato \n\n \n",
"bugtrack_url": null,
"license": null,
"summary": "MSA visualization python package for sequence analysis",
"version": "0.5.0",
"project_urls": {
"homepage": "https://moshi4.github.io/pyMSAviz/",
"repository": "https://github.com/moshi4/pyMSAviz/"
},
"split_keywords": [
"bioinformatics",
" matplotlib",
" sequence-alignment",
" sequence-analysis",
" visualization"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "19538760a22c39d7efcb040bd28042d6e1c79120ab2c0a8e81763ed8a1fd1eb7",
"md5": "72e8fd6a0f5db572dd12b3e158c3740e",
"sha256": "21645de22018d6592869d7e6de1b6f932f93681888b4035d6c830caafba57c9f"
},
"downloads": -1,
"filename": "pymsaviz-0.5.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "72e8fd6a0f5db572dd12b3e158c3740e",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.9",
"size": 17235,
"upload_time": "2024-09-14T03:25:49",
"upload_time_iso_8601": "2024-09-14T03:25:49.814748Z",
"url": "https://files.pythonhosted.org/packages/19/53/8760a22c39d7efcb040bd28042d6e1c79120ab2c0a8e81763ed8a1fd1eb7/pymsaviz-0.5.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "f055a7ffe31c8d389f5eed61713b226abb0a5f57b55ffd4e4c686b581b54f28c",
"md5": "eb7273ecef763ce040e5d7e5ac616e1a",
"sha256": "644a5bacfc0aa75b7c5d2c2e2b666020ac4f72e18b54a142b56d4638cfbe6764"
},
"downloads": -1,
"filename": "pymsaviz-0.5.0.tar.gz",
"has_sig": false,
"md5_digest": "eb7273ecef763ce040e5d7e5ac616e1a",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.9",
"size": 2005164,
"upload_time": "2024-09-14T03:25:51",
"upload_time_iso_8601": "2024-09-14T03:25:51.275597Z",
"url": "https://files.pythonhosted.org/packages/f0/55/a7ffe31c8d389f5eed61713b226abb0a5f57b55ffd4e4c686b581b54f28c/pymsaviz-0.5.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-09-14 03:25:51",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "moshi4",
"github_project": "pyMSAviz",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "pymsaviz"
}
