# pyranges
## Introduction
Pyranges is a Python library with a Rust backend for efficient and intuitive manipulation of genomics data,
particularly genomic intervals (like genes, genomic features, or reads).
The library is optimized for fast querying and manipulation of genomic annotations.
It enables intuitive and highly efficient pipelines for genomic analysis.
*"Finally ... This was what Python badly needed for years."* - Heng Li
## Version 1.x
This is version 1.x of pyranges. It is a complete rewrite of the original pyranges library,
that will replace the "default" (version 0) at the end in 2025. If you are a v0 user, check the migration guide
in the documentation.
## Documentation
The pyranges documentation, including installation instructions, API, tutorial, and how-to-pages, is
available at https://pyranges1.readthedocs.io/
## Install
Pyranges 1.x requires python ≥3.12. Minimal installation:
```bash
pip install pyranges1
```
Installation including all optional dependencies:
```bash
pip install pyranges1[all]
```
Details at https://pyranges1.readthedocs.io/en/latest/installation.html
## Features
- fast
- memory-efficient
- featureful
- pythonic/pandastic
## Paper/Cite
Stovner EB, Sætrom P (2020) PyRanges: efficient comparison of genomic intervals in Python.
*Bioinformatics 36(3):918-919* http://dx.doi.org/10.1093/bioinformatics/btz615
Coming soon: a paper for v1!
## Supporting pyranges
- most importantly, cite pyranges if you use it. It is the main metric funding sources care about.
- use pyranges in Stack Overflow/biostars questions and answers
- star the repo (possibly important for github visibility and as a proxy for project popularity)
## Asking for help
If you encounter bugs, or the documentation is not enough a cannot accomplish a specific task of interest,
or if you'd like new features implemented, open an Issue at github: https://github.com/pyranges/pyranges/issues
## Contributing to pyranges
Pyranges accepts code contributions in form of pull request.
For details, visit https://pyranges1.readthedocs.io/developer_guide.html
## Cheatsheet
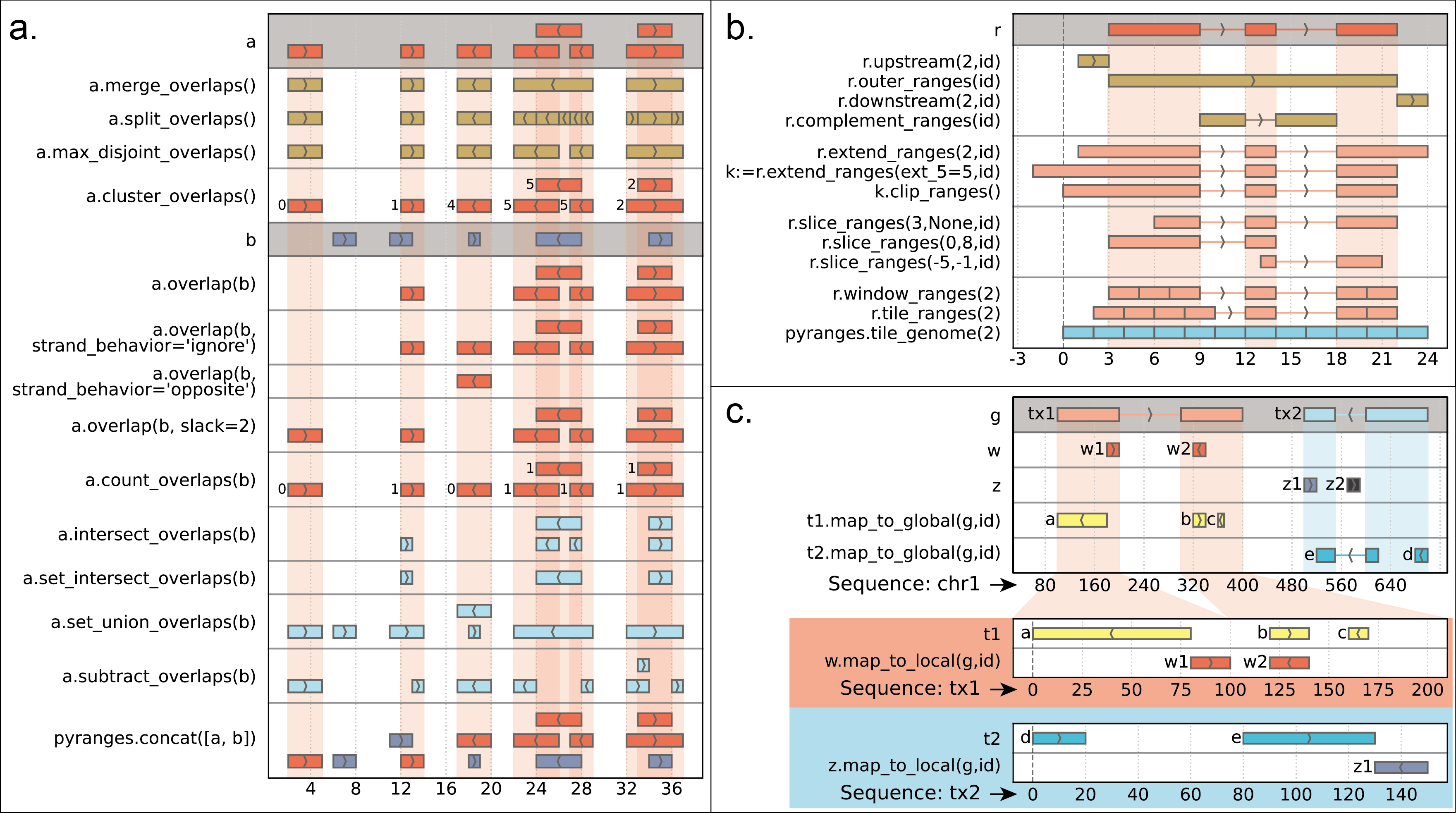
(The cheatsheet above was created with pyrangeyes, a companion graphical library: https://pyrangeyes.readthedocs.io/)
Raw data
{
"_id": null,
"home_page": null,
"name": "pyranges1",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.12.0",
"maintainer_email": null,
"keywords": "bioinformatics, genomicranges, genomics",
"author": null,
"author_email": "Endre Bakken Stovner <endbak@pm.me>, Marco Mariotti <marco.mariotti@ub.edu>",
"download_url": "https://files.pythonhosted.org/packages/ad/4e/9e3d1a8945a6fe826628d066b4fccbaec1816b611d151cdd2ba607879161/pyranges1-1.1.5.tar.gz",
"platform": null,
"description": "# pyranges\n\n## Introduction\n\nPyranges is a Python library with a Rust backend for efficient and intuitive manipulation of genomics data,\nparticularly genomic intervals (like genes, genomic features, or reads).\nThe library is optimized for fast querying and manipulation of genomic annotations.\nIt enables intuitive and highly efficient pipelines for genomic analysis.\n\n*\"Finally ... This was what Python badly needed for years.\"* - Heng Li\n\n## Version 1.x\nThis is version 1.x of pyranges. It is a complete rewrite of the original pyranges library, \nthat will replace the \"default\" (version 0) at the end in 2025. If you are a v0 user, check the migration guide \nin the documentation.\n\n## Documentation\n\nThe pyranges documentation, including installation instructions, API, tutorial, and how-to-pages, is \navailable at https://pyranges1.readthedocs.io/\n\n## Install\n\nPyranges 1.x requires python \u22653.12. Minimal installation: \n\n```bash\npip install pyranges1\n```\n\nInstallation including all optional dependencies:\n\n```bash\npip install pyranges1[all]\n```\n\nDetails at https://pyranges1.readthedocs.io/en/latest/installation.html\n\n\n## Features\n\n - fast\n - memory-efficient\n - featureful\n - pythonic/pandastic\n\n## Paper/Cite\n\nStovner EB, S\u00e6trom P (2020) PyRanges: efficient comparison of genomic intervals in Python. \n*Bioinformatics 36(3):918-919* http://dx.doi.org/10.1093/bioinformatics/btz615\n\nComing soon: a paper for v1!\n\n## Supporting pyranges\n\n - most importantly, cite pyranges if you use it. It is the main metric funding sources care about.\n - use pyranges in Stack Overflow/biostars questions and answers\n - star the repo (possibly important for github visibility and as a proxy for project popularity)\n\n## Asking for help\n\nIf you encounter bugs, or the documentation is not enough a cannot accomplish a specific task of interest, \nor if you'd like new features implemented, open an Issue at github: https://github.com/pyranges/pyranges/issues\n\n## Contributing to pyranges\n\nPyranges accepts code contributions in form of pull request. \nFor details, visit https://pyranges1.readthedocs.io/developer_guide.html\n\n## Cheatsheet\n\n(The cheatsheet above was created with pyrangeyes, a companion graphical library: https://pyrangeyes.readthedocs.io/)\n\n\n\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "GenomicRanges for Python.",
"version": "1.1.5",
"project_urls": {
"Homepage": "http://github.com/pyranges/pyranges_1.x"
},
"split_keywords": [
"bioinformatics",
" genomicranges",
" genomics"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "5430e5b49eb33e05da6f93623142555fc33389cb72778f0fbf4ebee52d2f9663",
"md5": "a248b534dead4294f596cda7b9586ae0",
"sha256": "de802aca602e89e49e023aafc1b93032d30b60f129765d1f74188aa5a5df8b05"
},
"downloads": -1,
"filename": "pyranges1-1.1.5-py3-none-any.whl",
"has_sig": false,
"md5_digest": "a248b534dead4294f596cda7b9586ae0",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.12.0",
"size": 937603,
"upload_time": "2025-10-19T15:11:31",
"upload_time_iso_8601": "2025-10-19T15:11:31.429165Z",
"url": "https://files.pythonhosted.org/packages/54/30/e5b49eb33e05da6f93623142555fc33389cb72778f0fbf4ebee52d2f9663/pyranges1-1.1.5-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "ad4e9e3d1a8945a6fe826628d066b4fccbaec1816b611d151cdd2ba607879161",
"md5": "052077f55f7d21981fd1689f17e79308",
"sha256": "9520ab3b9b4c06cf61939ed3cf8551973b44c11a3b69f6471ff137cea49ea6f3"
},
"downloads": -1,
"filename": "pyranges1-1.1.5.tar.gz",
"has_sig": false,
"md5_digest": "052077f55f7d21981fd1689f17e79308",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.12.0",
"size": 908594,
"upload_time": "2025-10-19T15:11:33",
"upload_time_iso_8601": "2025-10-19T15:11:33.169188Z",
"url": "https://files.pythonhosted.org/packages/ad/4e/9e3d1a8945a6fe826628d066b4fccbaec1816b611d151cdd2ba607879161/pyranges1-1.1.5.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-10-19 15:11:33",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "pyranges",
"github_project": "pyranges_1.x",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "pyranges1"
}
