
# Hydrogen Bond Analysis Tool (HBAT) v2
A Python package to automate the analysis of potential hydrogen bonds and similar type of weak interactions like halogen bonds and non-canonical interactions in macromolecular structures, available in Brookhaven Protein Database (PDB) file format. HBAT uses a geometric approach to identify potential hydrogen bonds by analyzing distance and angular criteria between donor-hydrogen-acceptor triplets.












[](https://codecov.io/gh/abhishektiwari/hbat)
[&replace=%241&style=plastic&logo=googlescholar&label=Cited%20By&cacheSeconds=86400)](https://scholar.google.com/citations?view_op=view_citation&hl=en&user=Mb7eYKYAAAAJ&citation_for_view=Mb7eYKYAAAAJ:u-x6o8ySG0sC)
[](https://socket.dev/pypi/package/hbat/overview/2.2.11/py3-none-any-whl)
[](https://www.codefactor.io/repository/github/abhishektiwari/hbat/overview/main)
## Background
HBAT v2 is a modern Python re-implementation of the original Perl-based tool developed by [Abhishek Tiwari](https://www.abhishek-tiwari.com) and Sunil Kumar Panigrahi.
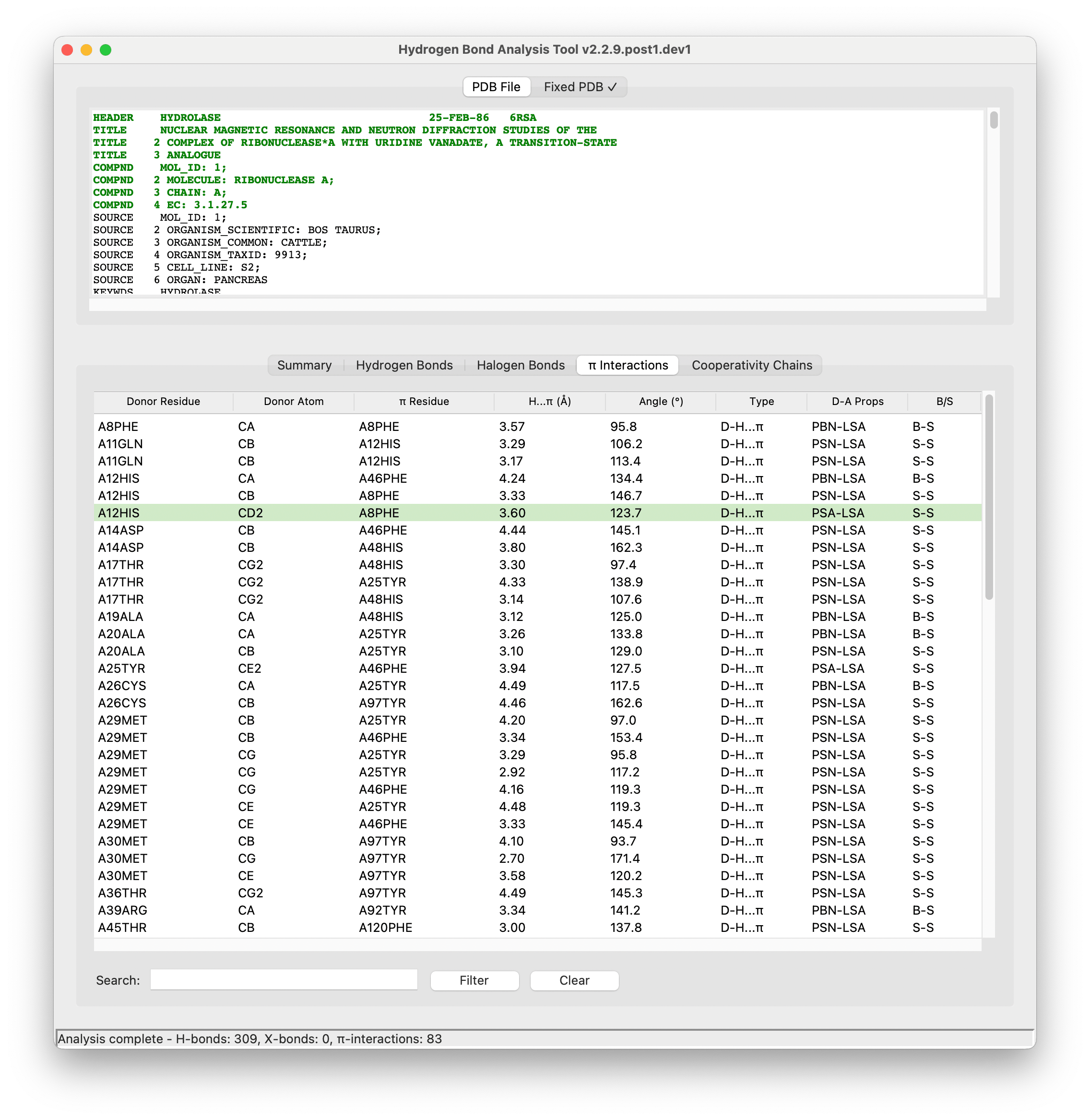
## Features
- **Comprehensive Analysis**: Detect and analyze potential hydrogen bonds, halogen bonds, and X-H...π interactions
- **Dual Interface**: Both graphical (tkinter) and command-line interfaces
- **Advanced Visualization**: Choice between NetworkX/matplotlib and GraphViz renderers for cooperativity chain visualization
- **High-Quality Graphics**: Export visualizations to PNG, SVG, PDF with configurable resolution
- **Interactive GUI**: Scrollable visualizations with dynamic layout switching and engine selection
- **Parameter Presets**: Built-in presets for different structure types (high-resolution, NMR, membrane proteins, etc.)
- **Flexible Parameters**: Customizable distance cutoffs, angle thresholds, and analysis modes.
- **Multiple Output Formats**: Text, CSV, and JSON export options
- **Fast Processing**: Optimized algorithms for efficient analysis of large structures
- **Cross-Platform**: Works on Windows, macOS, and Linux.
Please review [HBAT documentation](https://hbat.abhishek-tiwari.com/) for more details.
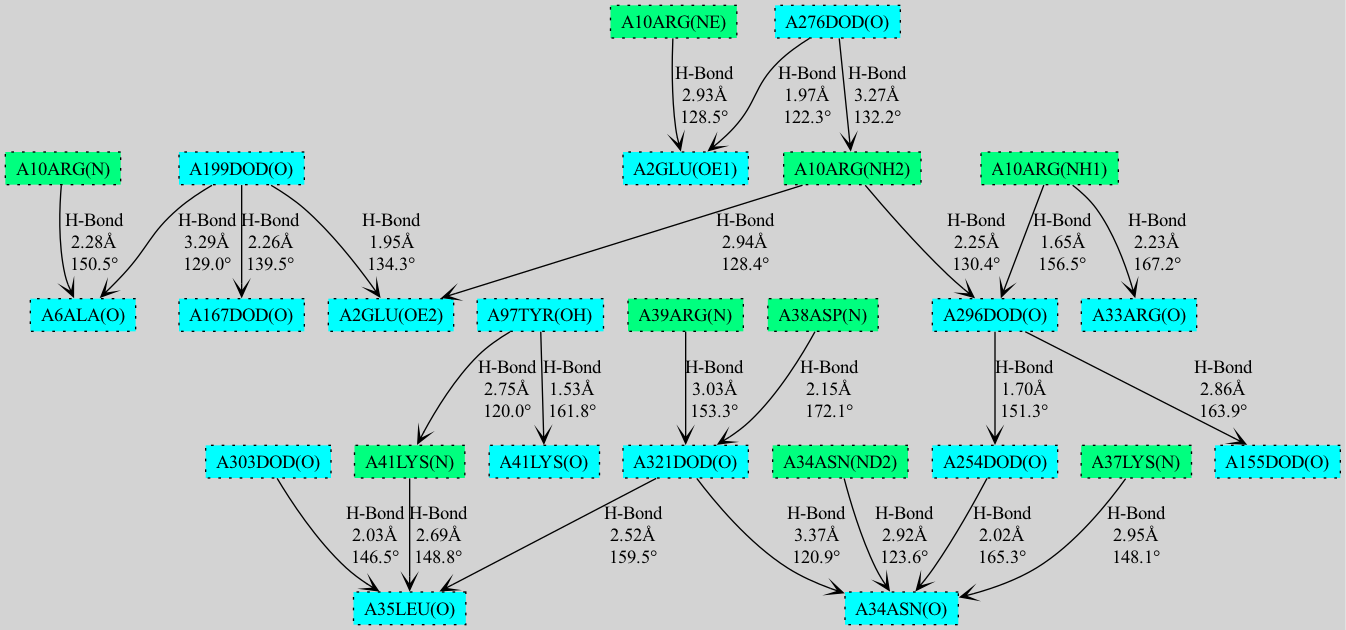
### Supported Interactions
1. **Hydrogen Bonds**: O-H...O, N-H...O, N-H...N, and other X-H...Y interactions
2. **Halogen Bonds**: C-X...Y interactions (X = F, Cl, Br, I; Y = N, O, S)
3. **X-H...π Interactions**: Hydrogen bonds to aromatic ring systems
Please review [HBAT documentation](https://hbat.abhishek-tiwari.com/) for more details.
## Installation
### Option 1: Install from PyPI (Recommended)
```bash
pip install hbat
```
Run HBAT Command-Line Interface (CLI) using `hbat` or launch HBAT GUI using `hbat-gui`.
### Option 2: Install from Source
```bash
git clone https://github.com/abhishektiwari/hbat.git
cd hbat
pip install -e .
```
Alternatively,
```bash
pip install git+https://github.com/abhishektiwari/hbat.git
```
Run HBAT Command-Line Interface (CLI) using `hbat` or launch HBAT GUI using `hbat-gui`.
### Option 3: Install from Conda
```
conda install -c hbat hbat
```
### Requirements
#### System Requirements
- **Python**: 3.9 or higher
- **tkinter**: tkinter is included with Python standard library on most systems. However, on Mac install Python and tkinter using `brew`.
```
brew install python python3-tk
```
- **GraphViz** (Optional): Required for advanced cooperativity chain visualization with high-quality graph rendering. HBAT will automatically fall back to NetworkX/matplotlib visualization if GraphViz is not available.
**Install GraphViz:**
On **Ubuntu/Debian**:
```bash
sudo apt-get update
sudo apt-get install graphviz
```
On **macOS** (using Homebrew):
```bash
brew install graphviz
```
On **Windows**:
- Download and install from [GraphViz official website](https://graphviz.org/download/)
- Or using Chocolatey: `choco install graphviz`
- Or using conda: `conda install -c conda-forge graphviz`
**Note**: After installing GraphViz, restart your terminal/command prompt before running HBAT to ensure the GraphViz executables are available in your PATH.
## Usage
### Graphical Interface
Launch the GUI application:
```bash
hbat-gui
```
The GUI provides,
- File browser for loading PDB files
- Parameter configuration panels
- Tabbed results display
- Export and visualization options
### Command-Line Interface
Basic usage:
```bash
hbat input.pdb
```
#### Output Format Options
HBAT supports multiple output formats with automatic detection based on file extension:
```bash
# Single file outputs (format auto-detected from extension)
hbat input.pdb -o results.txt # Text format
hbat input.pdb -o results.csv # CSV format (single file with all data)
hbat input.pdb -o results.json # JSON format (single file with all data)
# Multiple file outputs (separate files per interaction type)
hbat input.pdb --csv results # Creates results_h_bonds.csv, results_x_bonds.csv, etc.
hbat input.pdb --json results # Creates results_h_bonds.json, results_x_bonds.json, etc.
```
With custom parameters:
```bash
hbat input.pdb -o results.csv --hb-distance 3.0 --mode local
```
#### List Available Presets
```bash
hbat --list-presets
```
#### Use a specific preset
```bash
hbat protein.pdb --preset high_resolution
hbat membrane_protein.pdb --preset membrane_proteins
```
#### Use preset with custom overrides
```bash
hbat protein.pdb --preset drug_design_strict --hb-distance 3.0 --verbose
```
#### CLI Options
```
positional arguments:
input Input PDB file
optional arguments:
-h, --help show this help message and exit
-o OUTPUT, --output OUTPUT
Output file (format auto-detected from extension: .txt, .csv, .json)
--json JSON Export to multiple JSON files (base name for files)
--csv CSV Export to multiple CSV files (base name for files)
Preset Options:
--preset PRESET Load parameters from preset file (.hbat or .json)
--list-presets List available example presets and exit
Analysis Parameters:
--hb-distance HB_DISTANCE
Hydrogen bond H...A distance cutoff in Å (default: 3.5)
--hb-angle HB_ANGLE Hydrogen bond D-H...A angle cutoff in degrees (default: 120)
--da-distance DA_DISTANCE
Donor-acceptor distance cutoff in Å (default: 4.0)
--xb-distance XB_DISTANCE
Halogen bond X...A distance cutoff in Å (default: 4.0)
--xb-angle XB_ANGLE Halogen bond C-X...A angle cutoff in degrees (default: 120)
--pi-distance PI_DISTANCE
π interaction H...π distance cutoff in Å (default: 4.5)
--pi-angle PI_ANGLE π interaction D-H...π angle cutoff in degrees (default: 90)
--covalent-factor COVALENT_FACTOR
Covalent bond detection factor (default: 1.2)
--mode {complete,local}
Analysis mode: complete (all interactions) or local (intra-residue only)
Output Control:
--verbose, -v Verbose output with detailed progress
--quiet, -q Quiet mode with minimal output
--summary-only Output summary statistics only
Analysis Filters:
--no-hydrogen-bonds Skip hydrogen bond analysis
--no-halogen-bonds Skip halogen bond analysis
--no-pi-interactions Skip π interaction analysis
```
## License
This project is licensed under the MIT License - see the [LICENSE](LICENSE) file for details.
## Citation
If you use HBAT in your research, please cite:
```
@software{tiwari2025hbat,
author = {Tiwari, Abhishek},
title = {HBAT: Hydrogen Bond Analysis Tool},
version = {v2},
year = {2025},
url = {https://github.com/abhishektiwari/hbat}
}
```
```
@article{tiwari2007hbat,
author = {Tiwari, Abhishek and Panigrahi, Sunil Kumar},
doi = {10.3233/ISI-2007-00337},
journal = {In Silico Biology},
month = dec,
number = {6},
title = {{HBAT: A Complete Package for Analysing Strong and Weak Hydrogen Bonds in Macromolecular Crystal Structures}},
volume = {7},
year = {2007}
}
```
## Contributing
See our [contributing guide](CONTRIBUTING.md) and [development guide](https://hbat.abhishek-tiwari.com/development). At a high-level,
1. Fork the repository
2. Create a feature branch
3. Make your changes
4. Add tests if applicable
5. Submit a pull request
Raw data
{
"_id": null,
"home_page": null,
"name": "hbat",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": null,
"keywords": "bioinformatics, chemistry, hydrogen bonds, halogen bonds, protein structure, PDB, mmCIF",
"author": null,
"author_email": "Abhishek Tiwari <hbat@abhishek-tiwari.com>",
"download_url": "https://files.pythonhosted.org/packages/a0/18/062962f81fb8722c8e500bcf33d4d5c657aba989a4f36eb1e0ee2f70ecec/hbat-2.2.14.tar.gz",
"platform": null,
"description": "\n\n# Hydrogen Bond Analysis Tool (HBAT) v2 \n\nA Python package to automate the analysis of potential hydrogen bonds and similar type of weak interactions like halogen bonds and non-canonical interactions in macromolecular structures, available in Brookhaven Protein Database (PDB) file format. HBAT uses a geometric approach to identify potential hydrogen bonds by analyzing distance and angular criteria between donor-hydrogen-acceptor triplets.\n\n\n\n\n\n\n\n\n\n\n\n\n\n[](https://codecov.io/gh/abhishektiwari/hbat)\n[&replace=%241&style=plastic&logo=googlescholar&label=Cited%20By&cacheSeconds=86400)](https://scholar.google.com/citations?view_op=view_citation&hl=en&user=Mb7eYKYAAAAJ&citation_for_view=Mb7eYKYAAAAJ:u-x6o8ySG0sC)\n[](https://socket.dev/pypi/package/hbat/overview/2.2.11/py3-none-any-whl)\n[](https://www.codefactor.io/repository/github/abhishektiwari/hbat/overview/main)\n\n## Background\n\nHBAT v2 is a modern Python re-implementation of the original Perl-based tool developed by [Abhishek Tiwari](https://www.abhishek-tiwari.com) and Sunil Kumar Panigrahi.\n\n\n\n## Features\n\n- **Comprehensive Analysis**: Detect and analyze potential hydrogen bonds, halogen bonds, and X-H...\u03c0 interactions\n- **Dual Interface**: Both graphical (tkinter) and command-line interfaces\n- **Advanced Visualization**: Choice between NetworkX/matplotlib and GraphViz renderers for cooperativity chain visualization\n- **High-Quality Graphics**: Export visualizations to PNG, SVG, PDF with configurable resolution\n- **Interactive GUI**: Scrollable visualizations with dynamic layout switching and engine selection\n- **Parameter Presets**: Built-in presets for different structure types (high-resolution, NMR, membrane proteins, etc.)\n- **Flexible Parameters**: Customizable distance cutoffs, angle thresholds, and analysis modes.\n- **Multiple Output Formats**: Text, CSV, and JSON export options\n- **Fast Processing**: Optimized algorithms for efficient analysis of large structures\n- **Cross-Platform**: Works on Windows, macOS, and Linux.\n\nPlease review [HBAT documentation](https://hbat.abhishek-tiwari.com/) for more details.\n\n\n\n### Supported Interactions\n\n1. **Hydrogen Bonds**: O-H...O, N-H...O, N-H...N, and other X-H...Y interactions\n2. **Halogen Bonds**: C-X...Y interactions (X = F, Cl, Br, I; Y = N, O, S)\n3. **X-H...\u03c0 Interactions**: Hydrogen bonds to aromatic ring systems\n\nPlease review [HBAT documentation](https://hbat.abhishek-tiwari.com/) for more details.\n\n## Installation\n\n### Option 1: Install from PyPI (Recommended)\n\n```bash\npip install hbat\n```\n\nRun HBAT Command-Line Interface (CLI) using `hbat` or launch HBAT GUI using `hbat-gui`.\n\n### Option 2: Install from Source\n\n```bash\ngit clone https://github.com/abhishektiwari/hbat.git\ncd hbat\npip install -e .\n```\n\nAlternatively, \n\n```bash\npip install git+https://github.com/abhishektiwari/hbat.git\n```\n\nRun HBAT Command-Line Interface (CLI) using `hbat` or launch HBAT GUI using `hbat-gui`.\n\n### Option 3: Install from Conda\n\n```\nconda install -c hbat hbat\n```\n\n### Requirements\n\n#### System Requirements\n- **Python**: 3.9 or higher\n- **tkinter**: tkinter is included with Python standard library on most systems. However, on Mac install Python and tkinter using `brew`. \n\n```\nbrew install python python3-tk\n```\n\n- **GraphViz** (Optional): Required for advanced cooperativity chain visualization with high-quality graph rendering. HBAT will automatically fall back to NetworkX/matplotlib visualization if GraphViz is not available.\n\n**Install GraphViz:**\n\nOn **Ubuntu/Debian**:\n```bash\nsudo apt-get update\nsudo apt-get install graphviz\n```\n\nOn **macOS** (using Homebrew):\n```bash\nbrew install graphviz\n```\n\nOn **Windows**:\n- Download and install from [GraphViz official website](https://graphviz.org/download/)\n- Or using Chocolatey: `choco install graphviz`\n- Or using conda: `conda install -c conda-forge graphviz`\n\n**Note**: After installing GraphViz, restart your terminal/command prompt before running HBAT to ensure the GraphViz executables are available in your PATH.\n\n## Usage\n\n### Graphical Interface\n\nLaunch the GUI application:\n\n```bash\nhbat-gui\n```\n\nThe GUI provides,\n- File browser for loading PDB files\n- Parameter configuration panels\n- Tabbed results display\n- Export and visualization options\n\n### Command-Line Interface\n\nBasic usage:\n\n```bash\nhbat input.pdb\n```\n\n#### Output Format Options\n\nHBAT supports multiple output formats with automatic detection based on file extension:\n\n```bash\n# Single file outputs (format auto-detected from extension)\nhbat input.pdb -o results.txt # Text format\nhbat input.pdb -o results.csv # CSV format (single file with all data)\nhbat input.pdb -o results.json # JSON format (single file with all data)\n\n# Multiple file outputs (separate files per interaction type)\nhbat input.pdb --csv results # Creates results_h_bonds.csv, results_x_bonds.csv, etc.\nhbat input.pdb --json results # Creates results_h_bonds.json, results_x_bonds.json, etc.\n```\n\nWith custom parameters:\n\n```bash\nhbat input.pdb -o results.csv --hb-distance 3.0 --mode local\n```\n\n#### List Available Presets\n\n```bash\nhbat --list-presets\n```\n\n#### Use a specific preset\n\n```bash\nhbat protein.pdb --preset high_resolution\nhbat membrane_protein.pdb --preset membrane_proteins\n```\n\n#### Use preset with custom overrides\n\n```bash\nhbat protein.pdb --preset drug_design_strict --hb-distance 3.0 --verbose\n```\n\n#### CLI Options\n\n```\npositional arguments:\n input Input PDB file\n\noptional arguments:\n -h, --help show this help message and exit\n -o OUTPUT, --output OUTPUT\n Output file (format auto-detected from extension: .txt, .csv, .json)\n --json JSON Export to multiple JSON files (base name for files)\n --csv CSV Export to multiple CSV files (base name for files)\n\nPreset Options:\n --preset PRESET Load parameters from preset file (.hbat or .json)\n --list-presets List available example presets and exit\n\nAnalysis Parameters:\n --hb-distance HB_DISTANCE\n Hydrogen bond H...A distance cutoff in \u00c5 (default: 3.5)\n --hb-angle HB_ANGLE Hydrogen bond D-H...A angle cutoff in degrees (default: 120)\n --da-distance DA_DISTANCE\n Donor-acceptor distance cutoff in \u00c5 (default: 4.0)\n --xb-distance XB_DISTANCE\n Halogen bond X...A distance cutoff in \u00c5 (default: 4.0)\n --xb-angle XB_ANGLE Halogen bond C-X...A angle cutoff in degrees (default: 120)\n --pi-distance PI_DISTANCE\n \u03c0 interaction H...\u03c0 distance cutoff in \u00c5 (default: 4.5)\n --pi-angle PI_ANGLE \u03c0 interaction D-H...\u03c0 angle cutoff in degrees (default: 90)\n --covalent-factor COVALENT_FACTOR\n Covalent bond detection factor (default: 1.2)\n --mode {complete,local}\n Analysis mode: complete (all interactions) or local (intra-residue only)\n\nOutput Control:\n --verbose, -v Verbose output with detailed progress\n --quiet, -q Quiet mode with minimal output\n --summary-only Output summary statistics only\n\nAnalysis Filters:\n --no-hydrogen-bonds Skip hydrogen bond analysis\n --no-halogen-bonds Skip halogen bond analysis\n --no-pi-interactions Skip \u03c0 interaction analysis\n```\n\n## License\n\nThis project is licensed under the MIT License - see the [LICENSE](LICENSE) file for details.\n\n## Citation\n\nIf you use HBAT in your research, please cite:\n\n```\n@software{tiwari2025hbat,\n author = {Tiwari, Abhishek},\n title = {HBAT: Hydrogen Bond Analysis Tool},\n version = {v2},\n year = {2025},\n url = {https://github.com/abhishektiwari/hbat}\n}\n```\n\n```\n@article{tiwari2007hbat,\nauthor = {Tiwari, Abhishek and Panigrahi, Sunil Kumar},\ndoi = {10.3233/ISI-2007-00337},\njournal = {In Silico Biology},\nmonth = dec,\nnumber = {6},\ntitle = {{HBAT: A Complete Package for Analysing Strong and Weak Hydrogen Bonds in Macromolecular Crystal Structures}},\nvolume = {7},\nyear = {2007}\n}\n```\n\n## Contributing \n\nSee our [contributing guide](CONTRIBUTING.md) and [development guide](https://hbat.abhishek-tiwari.com/development). At a high-level,\n\n1. Fork the repository\n2. Create a feature branch\n3. Make your changes\n4. Add tests if applicable\n5. Submit a pull request\n",
"bugtrack_url": null,
"license": null,
"summary": "Hydrogen Bond Analysis Tool for PDB structures",
"version": "2.2.14",
"project_urls": {
"Bug Reports": "https://github.com/abhishektiwari/hbat/issues",
"Homepage": "https://github.com/abhishektiwari/hbat",
"Source": "https://github.com/abhishektiwari/hbat"
},
"split_keywords": [
"bioinformatics",
" chemistry",
" hydrogen bonds",
" halogen bonds",
" protein structure",
" pdb",
" mmcif"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "6a86e8e3fa9738485c2c1e3969f8dd6a0c41825061e08cad0dc1c51cf348a071",
"md5": "ff2e0f80198762fe345ae8f766a4e06a",
"sha256": "580c06a14c3a0f72dfd86df1bf05f37ef13c44e2d3012e28f0eeb91b497808a0"
},
"downloads": -1,
"filename": "hbat-2.2.14-py3-none-any.whl",
"has_sig": false,
"md5_digest": "ff2e0f80198762fe345ae8f766a4e06a",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 134907,
"upload_time": "2025-08-03T23:07:30",
"upload_time_iso_8601": "2025-08-03T23:07:30.914752Z",
"url": "https://files.pythonhosted.org/packages/6a/86/e8e3fa9738485c2c1e3969f8dd6a0c41825061e08cad0dc1c51cf348a071/hbat-2.2.14-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "a018062962f81fb8722c8e500bcf33d4d5c657aba989a4f36eb1e0ee2f70ecec",
"md5": "7a61ae124b910cf3d6ceb2ed60365556",
"sha256": "8efe103edb28efb40bd95d703f7bbd48c276f36d8a49a95038daaad9a3fde59c"
},
"downloads": -1,
"filename": "hbat-2.2.14.tar.gz",
"has_sig": false,
"md5_digest": "7a61ae124b910cf3d6ceb2ed60365556",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 2086188,
"upload_time": "2025-08-03T23:07:32",
"upload_time_iso_8601": "2025-08-03T23:07:32.610323Z",
"url": "https://files.pythonhosted.org/packages/a0/18/062962f81fb8722c8e500bcf33d4d5c657aba989a4f36eb1e0ee2f70ecec/hbat-2.2.14.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-08-03 23:07:32",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "abhishektiwari",
"github_project": "hbat",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [
{
"name": "pdbreader",
"specs": [
[
">=",
"0.1.0"
]
]
},
{
"name": "numpy",
"specs": [
[
">=",
"1.20.0"
]
]
},
{
"name": "networkx",
"specs": [
[
">=",
"2.5.0"
]
]
},
{
"name": "matplotlib",
"specs": [
[
">=",
"3.3.0"
]
]
},
{
"name": "graphviz",
"specs": [
[
">=",
"0.20.0"
]
]
},
{
"name": "Pillow",
"specs": [
[
">=",
"8.0.0"
]
]
},
{
"name": "pdbfixer-wheel",
"specs": [
[
">=",
"1.11.0"
]
]
},
{
"name": "openbabel-wheel",
"specs": [
[
">=",
"3.1.1.22"
]
]
},
{
"name": "pybel",
"specs": [
[
">=",
"0.15.5"
]
]
},
{
"name": "mmcif-pdbx",
"specs": [
[
">=",
"2.0.1"
]
]
},
{
"name": "mmcif",
"specs": [
[
">=",
"0.92.0"
]
]
},
{
"name": "tkinter-async-execute",
"specs": [
[
">=",
"1.4.0"
]
]
}
],
"lcname": "hbat"
}
