
# Lakeview
[](https://badge.fury.io/py/lakeview)
[](https://github.com/jzhang-dev/lakeview/actions/workflows/run_pytest.yml)
[](https://github.com/jzhang-dev/lakeview/actions/workflows/type_check_with_mypy.yml)
Lakeview is a Python 3 library for creating publication-quality [IGV](https://software.broadinstitute.org/software/igv/)-style genomic visualizations. Lakeview is based on [Matplotlib](https://matplotlib.org/).
A quick example:
```py
# Import Lakeview
import lakeview as lv
# Load aligned segments in a selected region from a BAM file
painter = lv.SequenceAlignment.from_file(
"PacBio_HiFi.bam", region="chr14:105,660,000-105,780,000"
)
# Create an empty GenomeViewer with two tracks
gv = lv.GenomeViewer(tracks=2, figsize=(8, 5), height_ratios=(1, 4))
# Plot alignment pileup
painter.draw_pileup(
gv.axes[0], # Plot on the first track of the GenomeViewer
show_mismatches=False, # Do not highlight mismatched bases
)
# Plot aligned segments
painter.draw_alignment(
gv.axes[1], # Plot on the second track of the GenomeViewer
show_mismatches=False, # Do not highlight mismatched bases
sort_by="length", # Plot longer reads first
link_by="name", # Link primary and supplementary alignments of the same read
max_rows=30, # Only show the first 30 alignment rows
)
# Adjust x axis limits
gv.set_xlim(105_670_000, 105_777_000)
# Save the plot
gv.savefig("example.png")
```
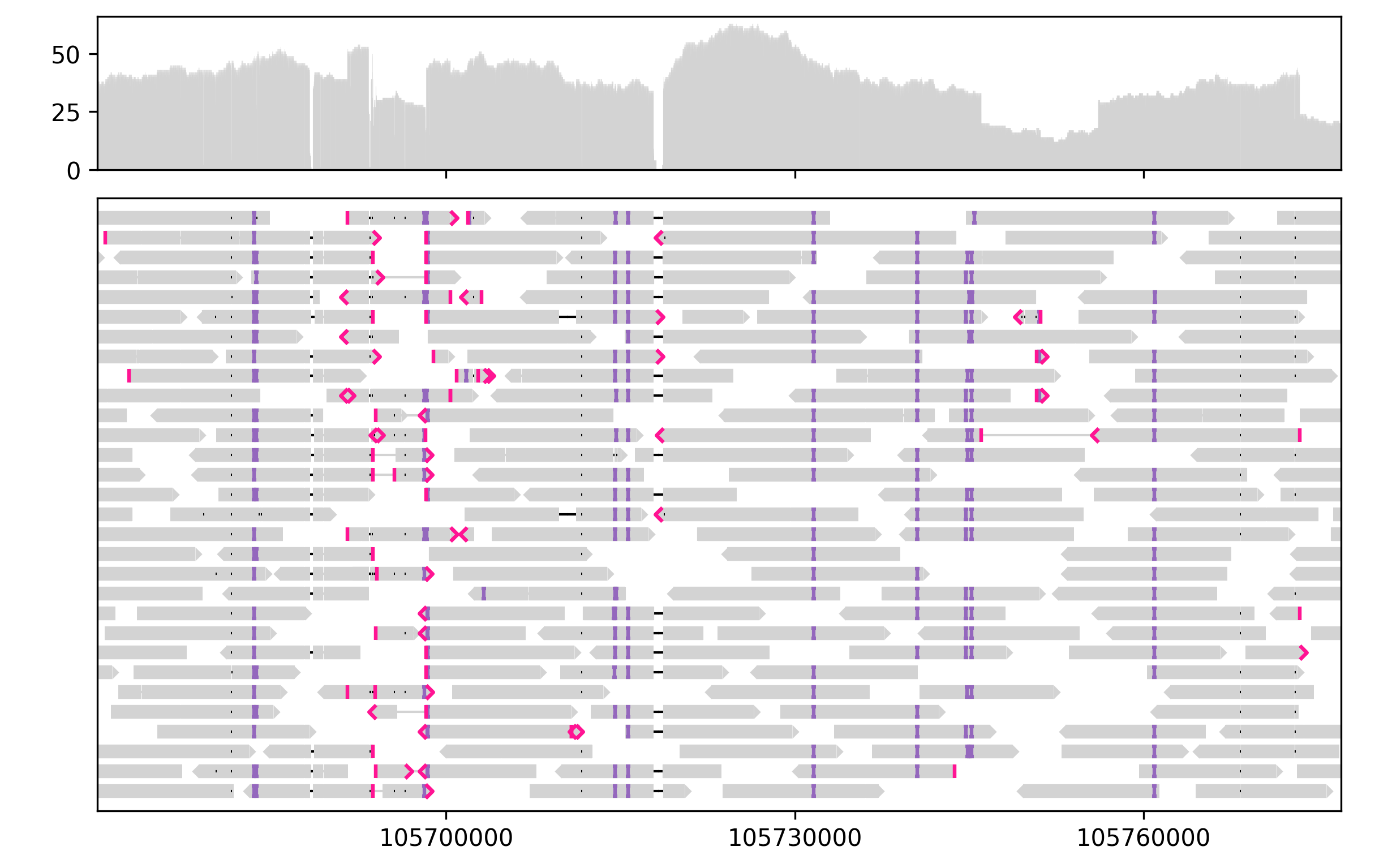
Lakeview currently supports visualzing sequence alignment, pileup, and gene annotation tracks. Additional functionality will be added in the future.
Try Lakeview interactively with Binder:
[](https://mybinder.org/v2/gh/jzhang-dev/lakeview/HEAD?labpath=docs%2Ftutorials%2Fquick_start.ipynb)
## Features
- **Improved clarity**. Lakeview inherits the familiar and intuitive visual style of [IGV](https://software.broadinstitute.org/software/igv/), with a clear layout designed for publication and presentation.
- **Programmable plotting**. Multiple files and genomic regions can be visualized automatically through a Pythonic interface inspired by [Seaborn](https://seaborn.pydata.org/) and [Pandas](https://pandas.pydata.org/).
- **Support for remote data**. Genomic data are often stored in remote servers without display devices. With Lakeview, you can plot remotely and view the output figures locally. Lakeview works well with [JupyterLab](https://jupyterlab.readthedocs.io/en/stable/) to streamline this workflow.
- **Transparency and reproducibility**. Figures are plotted transparently and annotated explicitly. The input data and the plotting code contain all the information needed to reproduce the figure.
- **Customizable layouts**. Lakeview supports many layouts implemented in [IGV](https://software.broadinstitute.org/software/igv/), while allowing the user to define custom rules for ordering, grouping, and coloring each segment. Advanced customization is possible via the [Matplotlib](https://matplotlib.org/) API.
- **Interactive plotting**. Lakeview ships with an interactive [Jupyter Widget](https://ipywidgets.readthedocs.io/en/stable/) to help users locate their regions of interest.
## Installation
```sh
pip install lakeview
```
## Documentation
Lakeview documentation is available at https://jzhang-dev.github.io/lakeview/.
## Contribute
Issues and pull requests are welcome via [GitHub](https://github.com/jzhang-dev/lakeview/).
A ready-to-use development environment is available via Gitpod:
[](https://gitpod.io/#https://github.com/jzhang-dev/lakeview)
## License
Lakeview is licensed under the GPL-3.0 license.
Raw data
{
"_id": null,
"home_page": "",
"name": "lakeview",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "",
"keywords": "bioinformatics,genomics,visualisation",
"author": "",
"author_email": "Jia-Yuan Zhang <jiayuan.zhang@ndm.ox.ac.uk>",
"download_url": "https://files.pythonhosted.org/packages/12/fb/8e6097ac1633e996782dd6069661d6f0ad3c3e66fc63689701be06f23d91/lakeview-0.2.0.tar.gz",
"platform": null,
"description": "\n\n# Lakeview\n\n[](https://badge.fury.io/py/lakeview)\n[](https://github.com/jzhang-dev/lakeview/actions/workflows/run_pytest.yml)\n[](https://github.com/jzhang-dev/lakeview/actions/workflows/type_check_with_mypy.yml)\n\nLakeview is a Python 3 library for creating publication-quality [IGV](https://software.broadinstitute.org/software/igv/)-style genomic visualizations. Lakeview is based on [Matplotlib](https://matplotlib.org/). \n\nA quick example:\n\n```py\n# Import Lakeview\nimport lakeview as lv\n\n# Load aligned segments in a selected region from a BAM file\npainter = lv.SequenceAlignment.from_file(\n \"PacBio_HiFi.bam\", region=\"chr14:105,660,000-105,780,000\"\n)\n# Create an empty GenomeViewer with two tracks\ngv = lv.GenomeViewer(tracks=2, figsize=(8, 5), height_ratios=(1, 4))\n# Plot alignment pileup\npainter.draw_pileup(\n gv.axes[0], # Plot on the first track of the GenomeViewer\n show_mismatches=False, # Do not highlight mismatched bases\n)\n# Plot aligned segments\npainter.draw_alignment(\n gv.axes[1], # Plot on the second track of the GenomeViewer\n show_mismatches=False, # Do not highlight mismatched bases\n sort_by=\"length\", # Plot longer reads first\n link_by=\"name\", # Link primary and supplementary alignments of the same read\n max_rows=30, # Only show the first 30 alignment rows\n)\n# Adjust x axis limits\ngv.set_xlim(105_670_000, 105_777_000)\n# Save the plot\ngv.savefig(\"example.png\")\n```\n\n\n\nLakeview currently supports visualzing sequence alignment, pileup, and gene annotation tracks. Additional functionality will be added in the future. \n\nTry Lakeview interactively with Binder:\n\n[](https://mybinder.org/v2/gh/jzhang-dev/lakeview/HEAD?labpath=docs%2Ftutorials%2Fquick_start.ipynb)\n \n\n## Features\n\n- **Improved clarity**. Lakeview inherits the familiar and intuitive visual style of [IGV](https://software.broadinstitute.org/software/igv/), with a clear layout designed for publication and presentation. \n- **Programmable plotting**. Multiple files and genomic regions can be visualized automatically through a Pythonic interface inspired by [Seaborn](https://seaborn.pydata.org/) and [Pandas](https://pandas.pydata.org/).\n- **Support for remote data**. Genomic data are often stored in remote servers without display devices. With Lakeview, you can plot remotely and view the output figures locally. Lakeview works well with [JupyterLab](https://jupyterlab.readthedocs.io/en/stable/) to streamline this workflow. \n- **Transparency and reproducibility**. Figures are plotted transparently and annotated explicitly. The input data and the plotting code contain all the information needed to reproduce the figure. \n- **Customizable layouts**. Lakeview supports many layouts implemented in [IGV](https://software.broadinstitute.org/software/igv/), while allowing the user to define custom rules for ordering, grouping, and coloring each segment. Advanced customization is possible via the [Matplotlib](https://matplotlib.org/) API.\n- **Interactive plotting**. Lakeview ships with an interactive [Jupyter Widget](https://ipywidgets.readthedocs.io/en/stable/) to help users locate their regions of interest.\n\n## Installation\n\n```sh\npip install lakeview\n```\n\n## Documentation\n\nLakeview documentation is available at https://jzhang-dev.github.io/lakeview/.\n\n## Contribute\n\nIssues and pull requests are welcome via [GitHub](https://github.com/jzhang-dev/lakeview/).\n\nA ready-to-use development environment is available via Gitpod:\n\n[](https://gitpod.io/#https://github.com/jzhang-dev/lakeview)\n\n## License\n\nLakeview is licensed under the GPL-3.0 license. \n\n",
"bugtrack_url": null,
"license": "GNU GENERAL PUBLIC LICENSE",
"summary": "A Python library for creating publication-quality genome visualisations.",
"version": "0.2.0",
"project_urls": {
"Bug Tracker": "https://github.com/jzhang-dev/lakeview/issues",
"Homepage": "https://jzhang-dev.github.io/lakeview/"
},
"split_keywords": [
"bioinformatics",
"genomics",
"visualisation"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "3e56e79dee560f6fdd7cf53db2bc7bb45bb047847330d947daa6737cfd8e289a",
"md5": "8b27f069747f361fb234b5db629333ab",
"sha256": "5137f68fdebf5fec8add5ddb54c7e1cfa7e9ca26e4857aeee0cbdc77145abb5e"
},
"downloads": -1,
"filename": "lakeview-0.2.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "8b27f069747f361fb234b5db629333ab",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 49299,
"upload_time": "2023-07-26T14:56:15",
"upload_time_iso_8601": "2023-07-26T14:56:15.150756Z",
"url": "https://files.pythonhosted.org/packages/3e/56/e79dee560f6fdd7cf53db2bc7bb45bb047847330d947daa6737cfd8e289a/lakeview-0.2.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "12fb8e6097ac1633e996782dd6069661d6f0ad3c3e66fc63689701be06f23d91",
"md5": "7f3e5ecaf6c23d96e3a88d47cb27db21",
"sha256": "e201adee252ea3c859f1c9f4efa71ba557fe5c0f8d789ecda7cef8aaa99c60cd"
},
"downloads": -1,
"filename": "lakeview-0.2.0.tar.gz",
"has_sig": false,
"md5_digest": "7f3e5ecaf6c23d96e3a88d47cb27db21",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 52187,
"upload_time": "2023-07-26T14:56:16",
"upload_time_iso_8601": "2023-07-26T14:56:16.176900Z",
"url": "https://files.pythonhosted.org/packages/12/fb/8e6097ac1633e996782dd6069661d6f0ad3c3e66fc63689701be06f23d91/lakeview-0.2.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-07-26 14:56:16",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "jzhang-dev",
"github_project": "lakeview",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "lakeview"
}
