# plotgen: easy data parsing and plot script generator
[](https://pypi.org/project/plotgen/)
[](https://github.com/bgottschall/plotgen/blob/master/LICENSE)
## About
Uses plotly to visualize data (e.g. tab, space, comma separated files) or pickled pandas dataframes. A lot of features are supported like reading in files with different separators, selecting and sorting columns, processing of multiple files, plotting line, scatter, bar, violin, box and gantt charts and customizing the graphs to all the needs. Many operations can be applied before plotting, like joining files, computing over rows and columns (sum, mean, median...), normalising data and filtering.
This script acts as a master script generating a plotting script. In case this script doesn't cover something that is required for a graph, it can output the final plotting script to make manual adjustments like adding annotations.
## Installation
Easy installation via pip:
```
pip install plotgen
```
### Manual
This is a single script project to allow the highest level of portability. Therefor manual installation is easy as you only have to put `plotgen` within your path and satisfy the requirements:
```
pip install -r requirements.txt
```
## Usage
The script is designed to process any formatted data file (e.g. tsv, csv, pandas df) into a table of rows and columns. Every row is a data point while every column is a data trace. Columns starting with '_' are treated as special columns (e.g. _error _labels, _colour or _offset). Rows starting with '#' are treated as comments and are ignored. (Those special characters can be adjusted with the rescpective parameters).
The input parsing arguments are processed in order and can be used as many times as whished. This enables very powerful preprocessing of the data before the actual plotting takes place. One can even do very complex inter-column and inter-row computation, normalisation, adding rows/columns with the average and a lot more. This script can also be used to just process the input data and output it as a new file or pickle container.
There are global parameters which can be set once per call like all colour options, legend options, width, height or output. The next set or parameters can only be specified on an input file and affect how the file is treated and plotted like row, col, sorting options, selection and ignoring of data points or plot type (line, bar, box, violin). However some options will be inherited by the following input files e.g. row, col and plot type and some won't be inherited like all input file parsing options (sorting, selection...) or options that affect the actual plot like axis titles or ranges. Options that are inherited can also be specified before an input file.
By default plotgen will create an html plot and opens it in the default browser:
```
plotgen -i lines.tsv
```
plotgen can also just print the data it has parsed from the file:
```
plotgen -i lines.tsv --print
```
Or create a standalone plotting script out of the input data which incorporates all the options that can be set and are shown later:
```
plotgen -i lines.tsv --output lines.py
```
All these options can also be combined to print the data, open a browser plot and save it as a PDF:
```
plotgen -i lines.tsv --print --browser --output lines.py lines.pdf --width 1920 --height 1080
```
The full power becomes visisble when working with multiple files (but also the same file more than once) and processing the data and making subplots:
```
plotgen -i lines.tsv --row 1 --select-columns Sin -i lines.tsv --row 2 --sort-order desc --sort-by-column Sin --select-column Cos
```
Selection of rows and columns is order sensitive and allows custom trace or data point ordering or duplication of traces and datapoints by repeating the names. All data row and column options also support numeric indexing by using e.g. --select-icolumns 0 1 2 etc.
The width and height of individual subplots can be simply adjusted by using colspan and rowspan:
```
plotgen -i lines.tsv --row 1 --rowspan 2 -i lines.tsv --row 3 --rowspan 1
```
It is also possible to plot many files into the same plot by just targetting the same row and col. Setting plot options like x-title, y-title or axis ranges should be done only once per subplot and ideally for the last file of the subplot.
## Examples
#### Line Chart
```
plotgen -i lines.tsv --plot line --output line.png
```

#### Scatter Chart
```
plotgen -i scatter.tsv --plot line --line-mode markers --output scatter.png
```
#### Bar Chart
```
plotgen -i bar.tsv --plot bar --output bar.png
```
#### Box Chart
```
plotgen -i distribution.tsv --plot box --output box.png
```
#### Violin Chart
```
plotgen -i distribution.tsv --plot violin --output violin.png
```
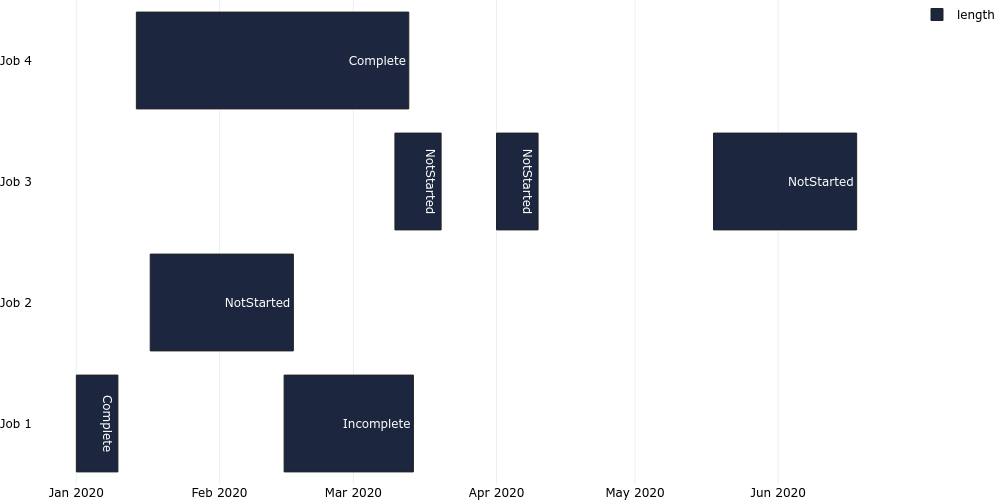
#### Gantt Chart
```
plotgen -i gantt.tsv --plot bar --bar-mode stack --bar-text-position inside --orientation h --x-type date --output gantt.png
```
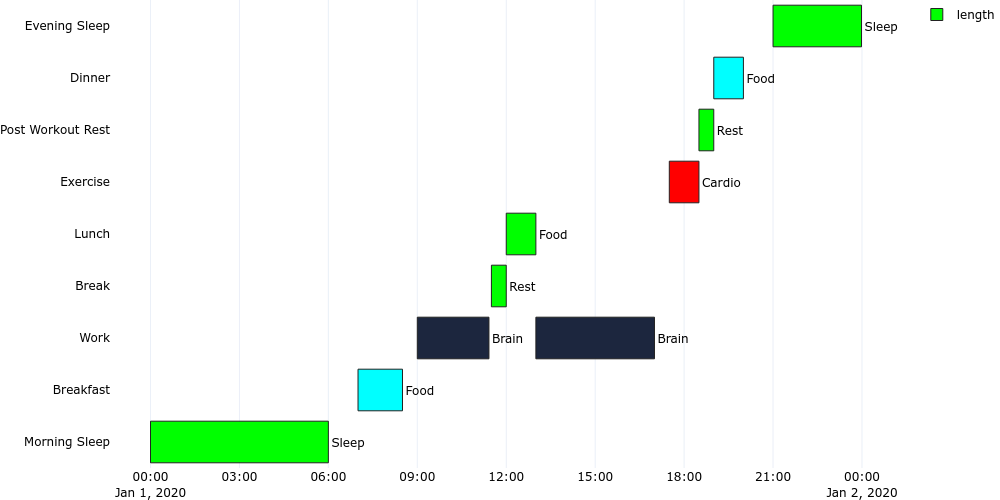
```
plotgen -i gantt_time.tsv --plot bar --bar-mode stack --bar-text-position outside --orientation h --x-type date --output gantt_time.png
```
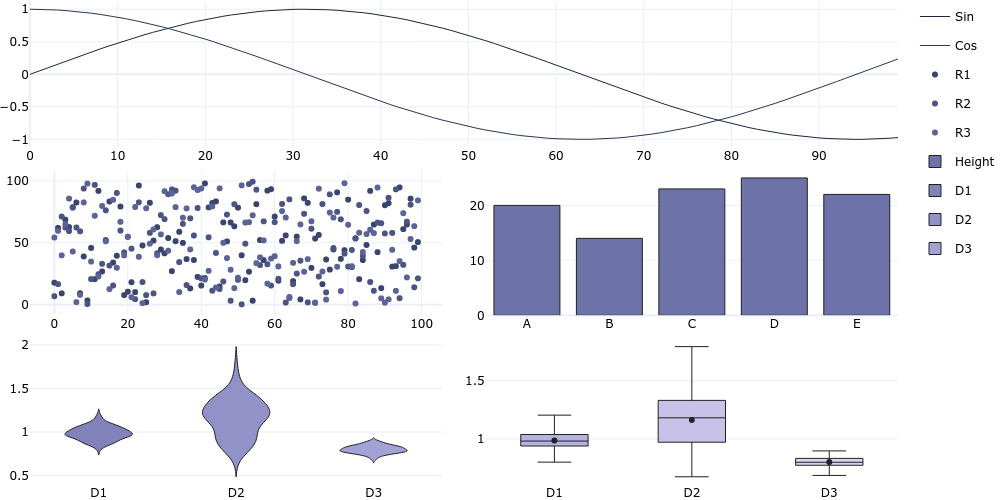
#### Combined Subplots
```
plotgen -i lines.tsv --colspan 2 -i scatter.tsv --row 2 --colspan 1 --line-mode markers -i bar.tsv --col 2 --plot bar -i distribution.csv --row 3 --col 1 --plot violin -i distribution.csv --col 2 --plot box --horizontal-spacing 0.05 --vertical-spacing 0.05 --per-trace-colours --output subplot.png
```
Raw data
{
"_id": null,
"home_page": "https://github.com/bgottschall/plotgen",
"name": "plotgen",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.9",
"maintainer_email": null,
"keywords": "tables graph plot parser plotly",
"author": "Bj\u00f6rn Gottschall",
"author_email": "info@gottschall.no",
"download_url": null,
"platform": null,
"description": "# plotgen: easy data parsing and plot script generator\n\n[](https://pypi.org/project/plotgen/)\n[](https://github.com/bgottschall/plotgen/blob/master/LICENSE)\n\n## About\n\nUses plotly to visualize data (e.g. tab, space, comma separated files) or pickled pandas dataframes. A lot of features are supported like reading in files with different separators, selecting and sorting columns, processing of multiple files, plotting line, scatter, bar, violin, box and gantt charts and customizing the graphs to all the needs. Many operations can be applied before plotting, like joining files, computing over rows and columns (sum, mean, median...), normalising data and filtering.\n\nThis script acts as a master script generating a plotting script. In case this script doesn't cover something that is required for a graph, it can output the final plotting script to make manual adjustments like adding annotations.\n\n## Installation\n\nEasy installation via pip:\n```\npip install plotgen\n```\n\n### Manual\n\nThis is a single script project to allow the highest level of portability. Therefor manual installation is easy as you only have to put `plotgen` within your path and satisfy the requirements:\n```\npip install -r requirements.txt\n```\n\n## Usage\n\nThe script is designed to process any formatted data file (e.g. tsv, csv, pandas df) into a table of rows and columns. Every row is a data point while every column is a data trace. Columns starting with '_' are treated as special columns (e.g. _error _labels, _colour or _offset). Rows starting with '#' are treated as comments and are ignored. (Those special characters can be adjusted with the rescpective parameters).\n\nThe input parsing arguments are processed in order and can be used as many times as whished. This enables very powerful preprocessing of the data before the actual plotting takes place. One can even do very complex inter-column and inter-row computation, normalisation, adding rows/columns with the average and a lot more. This script can also be used to just process the input data and output it as a new file or pickle container.\n\nThere are global parameters which can be set once per call like all colour options, legend options, width, height or output. The next set or parameters can only be specified on an input file and affect how the file is treated and plotted like row, col, sorting options, selection and ignoring of data points or plot type (line, bar, box, violin). However some options will be inherited by the following input files e.g. row, col and plot type and some won't be inherited like all input file parsing options (sorting, selection...) or options that affect the actual plot like axis titles or ranges. Options that are inherited can also be specified before an input file.\n\nBy default plotgen will create an html plot and opens it in the default browser:\n```\nplotgen -i lines.tsv\n```\n\nplotgen can also just print the data it has parsed from the file:\n```\nplotgen -i lines.tsv --print\n```\n\nOr create a standalone plotting script out of the input data which incorporates all the options that can be set and are shown later:\n```\nplotgen -i lines.tsv --output lines.py\n```\n\nAll these options can also be combined to print the data, open a browser plot and save it as a PDF:\n```\nplotgen -i lines.tsv --print --browser --output lines.py lines.pdf --width 1920 --height 1080\n```\n\n\nThe full power becomes visisble when working with multiple files (but also the same file more than once) and processing the data and making subplots:\n```\nplotgen -i lines.tsv --row 1 --select-columns Sin -i lines.tsv --row 2 --sort-order desc --sort-by-column Sin --select-column Cos \n```\nSelection of rows and columns is order sensitive and allows custom trace or data point ordering or duplication of traces and datapoints by repeating the names. All data row and column options also support numeric indexing by using e.g. --select-icolumns 0 1 2 etc.\n\nThe width and height of individual subplots can be simply adjusted by using colspan and rowspan:\n```\nplotgen -i lines.tsv --row 1 --rowspan 2 -i lines.tsv --row 3 --rowspan 1\n```\n\nIt is also possible to plot many files into the same plot by just targetting the same row and col. Setting plot options like x-title, y-title or axis ranges should be done only once per subplot and ideally for the last file of the subplot. \n\n## Examples\n\n#### Line Chart\n```\nplotgen -i lines.tsv --plot line --output line.png\n```\n\n#### Scatter Chart\n```\nplotgen -i scatter.tsv --plot line --line-mode markers --output scatter.png\n```\n\n#### Bar Chart\n```\nplotgen -i bar.tsv --plot bar --output bar.png\n```\n\n#### Box Chart\n```\nplotgen -i distribution.tsv --plot box --output box.png\n```\n\n#### Violin Chart\n```\nplotgen -i distribution.tsv --plot violin --output violin.png\n```\n\n#### Gantt Chart\n```\nplotgen -i gantt.tsv --plot bar --bar-mode stack --bar-text-position inside --orientation h --x-type date --output gantt.png\n```\n\n```\nplotgen -i gantt_time.tsv --plot bar --bar-mode stack --bar-text-position outside --orientation h --x-type date --output gantt_time.png\n```\n\n#### Combined Subplots\n```\nplotgen -i lines.tsv --colspan 2 -i scatter.tsv --row 2 --colspan 1 --line-mode markers -i bar.tsv --col 2 --plot bar -i distribution.csv --row 3 --col 1 --plot violin -i distribution.csv --col 2 --plot box --horizontal-spacing 0.05 --vertical-spacing 0.05 --per-trace-colours --output subplot.png\n```\n\n\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A plotly plotting script generator and data parser",
"version": "0.8.18",
"project_urls": {
"Homepage": "https://github.com/bgottschall/plotgen"
},
"split_keywords": [
"tables",
"graph",
"plot",
"parser",
"plotly"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "68695ca8bba53ec5a8c8105538c84baee1fe4c708f5ff8955de59c05e309070e",
"md5": "94b35fda5105d159a0de9454f442b9b8",
"sha256": "755fa7d64a924d26ad702740ec0209258c103436b4314594835b7db0df2838d9"
},
"downloads": -1,
"filename": "plotgen-0.8.18-py3-none-any.whl",
"has_sig": false,
"md5_digest": "94b35fda5105d159a0de9454f442b9b8",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.9",
"size": 40127,
"upload_time": "2024-04-29T07:25:29",
"upload_time_iso_8601": "2024-04-29T07:25:29.070585Z",
"url": "https://files.pythonhosted.org/packages/68/69/5ca8bba53ec5a8c8105538c84baee1fe4c708f5ff8955de59c05e309070e/plotgen-0.8.18-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-04-29 07:25:29",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "bgottschall",
"github_project": "plotgen",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"requirements": [
{
"name": "scikit-learn",
"specs": [
[
">=",
"1.3.2"
]
]
},
{
"name": "scipy",
"specs": [
[
">=",
"1.11.3"
]
]
},
{
"name": "argcomplete",
"specs": [
[
">=",
"3.1.4"
]
]
},
{
"name": "plotly",
"specs": [
[
">=",
"5.18.0"
]
]
},
{
"name": "numpy",
"specs": [
[
">=",
"1.26.1"
]
]
},
{
"name": "pandas",
"specs": [
[
">=",
"2.2.0"
]
]
},
{
"name": "xopen",
"specs": [
[
">=",
"1.8.0"
]
]
},
{
"name": "kaleido",
"specs": [
[
">=",
"0.2.1"
]
]
},
{
"name": "seaborn",
"specs": [
[
">=",
"0.13.0"
]
]
},
{
"name": "argcomplete",
"specs": [
[
">=",
"3.1.4"
]
]
},
{
"name": "pyarrow",
"specs": [
[
">=",
"15.0.0"
]
]
}
],
"lcname": "plotgen"
}
