<p align="center">
<img src="https://raw.githubusercontent.com/adaptyvbio/ProteinFlow/main/media/proteinflow_logo_small.png" width="60">
</p>
<p align="center">
<b> ProteinFlow - A data processing pipeline for all your protein design needs </b> <br />
</p>
<p align="center">
<a href="https://adaptyvbio.github.io/ProteinFlow/" target="_blank">
Docs
</a>
</p>
---
[](https://opensource.org/licenses/BSD-3-Clause)
[](https://github.com/psf/black)
[](https://pypi.org/project/proteinflow/)
[](https://anaconda.org/adaptyvbio/proteinflow)
[](https://hub.docker.com/r/adaptyvbio/proteinflow/tags)
[](CONTRIBUTING.md)
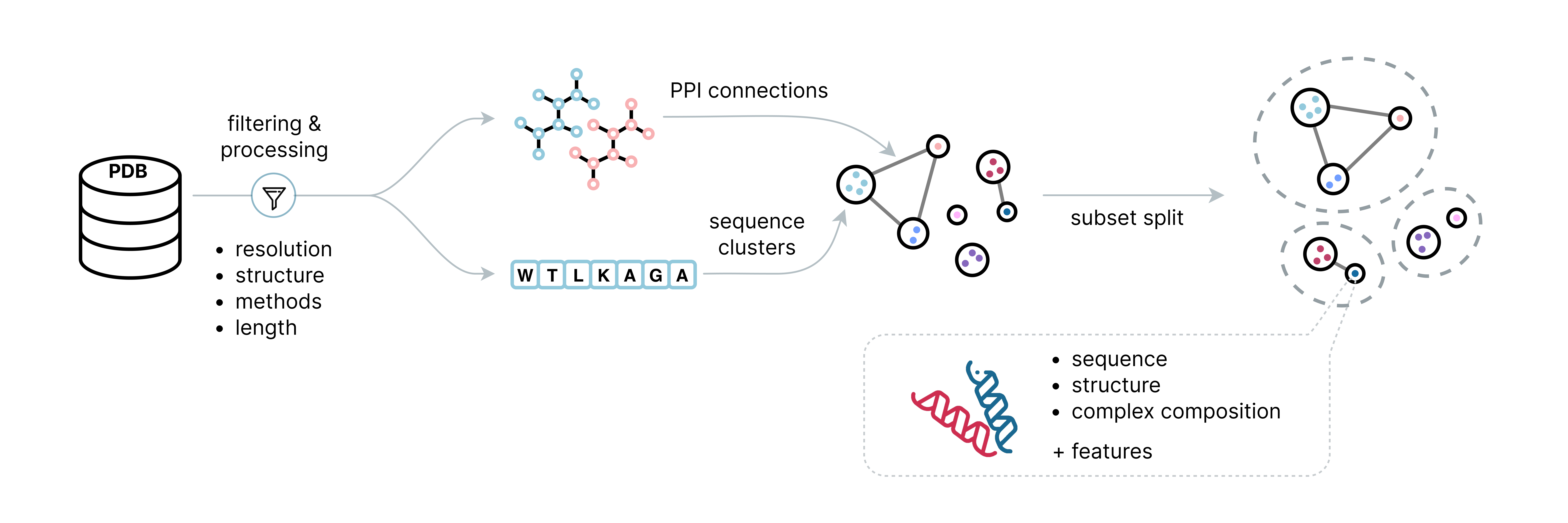
ProteinFlow is an open-source Python library that streamlines the pre-processing of protein structure data for deep learning applications. ProteinFlow enables users to efficiently filter, cluster, and generate new datasets from resources like the Protein Data Bank (PDB) and SAbDab (The Structural Antibody Database).
Here are some of the key features we currently support:
- ⛓️ Processing of both single-chain and multi-chain protein structures (Biounit PDB definition)
- 🏷️ Various featurization options can be computed, including secondary structure features, torsion angles, etc.
- 💾 A variety of data loading options and conversions to cater to different downstream training frameworks
- 🧬 Access to up-to-date, pre-computed protein structure datasets
---
## Installation
conda:
```bash
# This should take a few minutes, be patient
conda install -c conda-forge -c bioconda -c adaptyvbio proteinflow
```
pip:
```bash
pip install proteinflow
```
docker:
```bash
docker pull adaptyvbio/proteinflow
```
By default installing `proteinflow` with conda or pip will only load the dependencies that are required for the main functions of the package: downloading, generating and splitting datasets. If you are interested in using other functions like visualization, metrics and other data processing methods, please install the package with `pip install proteinflow[processing]` or use the docker image.
Some metric functions also have separate requirements, see the documentation for details. All of them are installed in the docker image.
### Troubleshooting
- If you are using python 3.10 and encountering installation problems, try running `python -m pip install prody==2.4.0` before installing `proteinflow`.
- If you are planning to generate new datasets and installed `proteinflow` with `pip` (or with `conda` on Mac OS with an M1 processor), you will need to additionally install [`mmseqs`](https://github.com/soedinglab/MMseqs2).
- Generating new datasets also depends on the `rcsbsearch` package and the latest release [v0.2.3](https://github.com/sbliven/rcsbsearch/releases/tag/v0.2.3) is currently not working correctly. The recommended fix is installing the version from [this pull request](https://github.com/sbliven/rcsbsearch/pull/6).
```bash
python -m pip install "rcsbsearch @ git+https://github.com/sbliven/rcsbsearch@dbdfe3880cc88b0ce57163987db613d579400c8e"
```
- The docker image can be accessed in interactive mode with this command.
```bash
docker run -it -v /path/to/data:/media adaptyvbio/proteinflow bash
```
## Usage
### Downloading pre-computed datasets (stable)
Already precomputed datasets with consensus set of parameters and can be accessed and downloaded using the `proteinflow`. package. Check the output of `proteinflow check_tags` for a list of available tags.
```bash
proteinflow download --tag 20230102_stable
```
### Running the pipeline (PDB)
You can also run `proteinflow` with your own parameters. Check the output of `proteinflow check_snapshots` for a list of available PDB snapshots (naming rule: `yyyymmdd`).
For instance, let's generate a dataset with the following description:
- resolution threshold: 5 angstrom,
- PDB snapshot: 20190101,
- structure methods accepted: all (x-ray christolography, NRM, Cryo-EM),
- sequence identity threshold for clustering: 40% sequence similarity,
- maximum length per sequence: 1000 residues,
- minimum length per sequence: 5 residues,
- maximum fraction of missing values at the ends: 10%,
- size of validation subset: 10%.
```bash
proteinflow generate --tag new --resolution_thr 5 --pdb_snapshot 20190101 --not_filter_methods --min_seq_id 0.4 --max_length 1000 --min_length 5 --missing_ends_thr 0.1 --valid_split 0.1
```
See the [docs](https://adaptyvbio.github.io/ProteinFlow/) (or `proteinflow generate --help`) for the full list of parameters and more information.
A registry of all the files that are removed during the filtering as well as description with the reason for their removal is created automatically for each `generate` command. The log files are save (at `data/logs` by default) and a summary can be accessed running `proteinflow get_summary {log_path}`.
### Running the pipeline (SAbDab)
You can also use the `--sabdab` option in `proteinflow generate` to load files from SAbDab and cluster them based on CDRs. By default the `--sabdab` tag will download the latest up-to-date version of the SabDab dataset and cluster the antibodies based on their CDR sequence.
Alternatively, it can be used together with the tag `--sabdab_data_path` to process a custom SAbDab-like zip file or folder. This allows you to use search and query tools from the [SabDab web interface](https://opig.stats.ox.ac.uk/webapps/newsabdab/sabdab/) to create a custom dataset by downloading the archived zip file of the structures selected. (Under Downloads section of your SabDab query).
SAbDab sequences clustering is done across all 6 Complementary Determining Regions (CDRs) - H1, H2, H3, L1, L2, L3, based on the [Chothia numbering](https://pubmed.ncbi.nlm.nih.gov/9367782/) implemented by SabDab. CDRs from nanobodies and other synthetic constructs are clustered together with other heavy chain CDRs. The resulting CDR clusters are split into training, test and validation in a way that ensures that every PDB file only appears in one subset.
Individual output pickle files represent heavy chain - light chain - antigen complexes (created from SAbDab annotation, sometimes more than one per PDB entry). Each of the elements (heavy chain, light chain, antigen) can be missing in specific entries and there can be multiple antigen chains. In order to filter for at least one antigen chain, use the `--require_antigen` option.
For instance, let's generate a dataset with the following description:
- SabDab version: latest (up-to-date),
- resolution threshold: 5 angstrom,
- structure methods accepted: all (x-ray christolography, NRM, Cryo-EM),
- sequence identity threshold for clustering (CDRs): 40%,
- size of validation subset: 10%.
```bash
proteinflow generate --sabdab --tag new --resolution_thr 5 --not_filter_methods --min_seq_id 0.4 --valid_split 0.1
```
### Splitting
By default, both `proteinflow generate` and `proteinflow download` will also split your data into training, test and validation according to MMseqs2 clustering and homomer/heteromer/single chain proportions. However, you can skip this step with a `--skip_splitting` flag and then perform it separately with the `proteinflow split` command.
The following command will perform the splitting with a 10% validation set, a 5% test set and a 50% threshold for sequence identity clusters.
```bash
proteinflow split --tag new --valid_split 0.1 --test_split 0.5 --min_seq_id 0.5
```
Use the `--exclude_chains` and `--exclude_threshold` parameters to move all biounits that contain chains similar to what you specify to a separate folder.
### Using the data
The output files are pickled nested dictionaries where first-level keys are chain Ids and second-level keys are the following:
- `'crd_bb'`: a `numpy` array of shape `(L, 4, 3)` with backbone atom coordinates (N, C, CA, O),
- `'crd_sc'`: a `numpy` array of shape `(L, 10, 3)` with sidechain atom coordinates (check `proteinflow.sidechain_order()` for the order of atoms),
- `'msk'`: a `numpy` array of shape `(L,)` where ones correspond to residues with known coordinates and
zeros to missing values,
- `'seq'`: a string of length `L` with residue types.
In a SAbDab datasets, an additional key is added to the dictionary:
- `'cdr'`: a `numpy` array of shape `(L,)` where CDR residues are marked with the corresponding type (`'H1'`, `'L1'`, ...)
and non-CDR residues are marked with `'-'`.
Note that the sequence information in the PDB files is aligned to the FASTA sequences to identify the missing residues.
Once your data is ready, you can open the files with `pickle` directly.
```python
import pickle
import os
train_folder = "./data/proteinflow_new/training"
for filename in os.listdir(train_folder):
with open(os.path.join(train_folder, filename), "rb") as f:
data = pickle.load(f)
crd_bb = data["crd_bb"]
seq = data["seq"]
...
```
Alternatively, you can use our `ProteinDataset` or `ProteinLoader` classes
for convenient processing. Among other things, they allow for feature extraction, single chain / homomer / heteromer filtering and randomized sampling from sequence identity clusters.
For example, here is how we can create a data loader that:
- samples a different cluster representative at every epoch,
- extracts dihedral angles, sidechain orientation and secondary structure features,
- only loads pairs of interacting proteins (larger biounits are broken up into pairs),
- has batch size 8.
```python
from proteinflow import ProteinLoader
train_loader = ProteinLoader.from_args(
"./data/proteinflow_new/training",
clustering_dict_path="./data/proteinflow_new/splits_dict/train.pickle",
node_features_type="dihedral+sidechain_orientation+secondary_structure",
entry_type="pair",
batch_size=8,
)
for batch in train_loader:
crd_bb = batch["X"] #(B, L, 4, 3)
seq = batch["S"] #(B, L)
sse = batch["secondary_structure"] #(B, L, 3)
to_predict = batch["masked_res"] #(B, L), 1 where the residues should be masked, 0 otherwise
...
```
See more details on available parameters and the data format in the [docs](https://adaptyvbio.github.io/ProteinFlow/) + [this repository](https://github.com/adaptyvbio/ProteinFlow-models) for a use case.
## ProteinFlow Stable Releases
You can download them with `proteinflow download --tag {tag}` in the command line or browse in the [interface](https://proteinflow-datasets.s3.eu-west-1.amazonaws.com/index.html).
|Tag |Date |Snapshot|Size|Min res|Min len|Max len|Max chains|MMseqs thr|Split (train/val/test)|Missing thr (ends/middle)|Source|Remove redundancies|Note|
|-------|--------|--------|----|-------|-------|-------|----------|----------|----------------------|-------------------------|---|---|----------------|
|paper|10.11.22|20220103|24G|3.5|30|10'000|-|0.3|90/5/5|0.3/0.1|PDB|yes|first release, no mmCIF files|
|20230102_stable|27.02.23|20230102|28G|3.5|30|10'000|-|0.3|90/5/5|0.3/0.1|PDB|yes|v1.1.1|
|20230623_sabdab|26.06.23|live 26.06.23|1.4G|3.5|30|10'000|-|0.3|96/3/1|0.5/0.2|SAbDab|no|v1.4.1 (requires >= v1.4.0)|
|20230102_v200|19.07.23|20230102|33G|3.5|30|10'000|10|0.3|90/5/5|0.3/0.1|PDB|no|v2.0.0|
|20231221_sabdab|21.12.23|live 21.12.23|1.8G|3.5|30|10'000|-|0.3|96/3/1|0.5/0.2|SAbDab|no|v2.6.1 (requires >= v1.4.0)|
## License
The `proteinflow` package and data are released and distributed under the BSD 3-Clause License
## Contributions
This is an open source project supported by [Adaptyv Bio](https://www.adaptyvbio.com/). Contributions, suggestions and bug-fixes are welcomed.
Raw data
{
"_id": null,
"home_page": "",
"name": "proteinflow",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "",
"keywords": "bioinformatics,dataset,protein,PDB,deep learning,antibody",
"author": "",
"author_email": "Liza Kozlova <liza@adaptyvbio.com>",
"download_url": "https://files.pythonhosted.org/packages/92/72/aab58ae3836bf1fd3abcf7788ba51a1b45e4d5e5d4640d6b0a69942a04d9/proteinflow-2.8.0.tar.gz",
"platform": null,
"description": "<p align=\"center\">\n <img src=\"https://raw.githubusercontent.com/adaptyvbio/ProteinFlow/main/media/proteinflow_logo_small.png\" width=\"60\">\n</p>\n\n<p align=\"center\">\n <b> ProteinFlow - A data processing pipeline for all your protein design needs </b> <br />\n</p>\n\n<p align=\"center\">\n <a href=\"https://adaptyvbio.github.io/ProteinFlow/\" target=\"_blank\">\n Docs\n </a>\n</p>\n\n---\n\n[](https://opensource.org/licenses/BSD-3-Clause)\n[](https://github.com/psf/black)\n[](https://pypi.org/project/proteinflow/)\n[](https://anaconda.org/adaptyvbio/proteinflow)\n[](https://hub.docker.com/r/adaptyvbio/proteinflow/tags)\n[](CONTRIBUTING.md)\n\n\nProteinFlow is an open-source Python library that streamlines the pre-processing of protein structure data for deep learning applications. ProteinFlow enables users to efficiently filter, cluster, and generate new datasets from resources like the Protein Data Bank (PDB) and SAbDab (The Structural Antibody Database).\n\nHere are some of the key features we currently support:\n\n- \u26d3\ufe0f Processing of both single-chain and multi-chain protein structures (Biounit PDB definition)\n- \ud83c\udff7\ufe0f Various featurization options can be computed, including secondary structure features, torsion angles, etc.\n- \ud83d\udcbe A variety of data loading options and conversions to cater to different downstream training frameworks\n- \ud83e\uddec Access to up-to-date, pre-computed protein structure datasets\n\n\n\n---\n\n## Installation\nconda:\n```bash\n# This should take a few minutes, be patient\nconda install -c conda-forge -c bioconda -c adaptyvbio proteinflow\n```\n\npip:\n```bash\npip install proteinflow\n```\n\ndocker:\n```bash\ndocker pull adaptyvbio/proteinflow\n```\n\nBy default installing `proteinflow` with conda or pip will only load the dependencies that are required for the main functions of the package: downloading, generating and splitting datasets. If you are interested in using other functions like visualization, metrics and other data processing methods, please install the package with `pip install proteinflow[processing]` or use the docker image.\n\nSome metric functions also have separate requirements, see the documentation for details. All of them are installed in the docker image.\n\n### Troubleshooting\n- If you are using python 3.10 and encountering installation problems, try running `python -m pip install prody==2.4.0` before installing `proteinflow`.\n- If you are planning to generate new datasets and installed `proteinflow` with `pip` (or with `conda` on Mac OS with an M1 processor), you will need to additionally install [`mmseqs`](https://github.com/soedinglab/MMseqs2).\n- Generating new datasets also depends on the `rcsbsearch` package and the latest release [v0.2.3](https://github.com/sbliven/rcsbsearch/releases/tag/v0.2.3) is currently not working correctly. The recommended fix is installing the version from [this pull request](https://github.com/sbliven/rcsbsearch/pull/6).\n```bash\npython -m pip install \"rcsbsearch @ git+https://github.com/sbliven/rcsbsearch@dbdfe3880cc88b0ce57163987db613d579400c8e\"\n```\n- The docker image can be accessed in interactive mode with this command.\n```bash\ndocker run -it -v /path/to/data:/media adaptyvbio/proteinflow bash\n```\n\n## Usage\n### Downloading pre-computed datasets (stable)\nAlready precomputed datasets with consensus set of parameters and can be accessed and downloaded using the `proteinflow`. package. Check the output of `proteinflow check_tags` for a list of available tags.\n```bash\nproteinflow download --tag 20230102_stable \n```\n\n### Running the pipeline (PDB)\nYou can also run `proteinflow` with your own parameters. Check the output of `proteinflow check_snapshots` for a list of available PDB snapshots (naming rule: `yyyymmdd`).\n\nFor instance, let's generate a dataset with the following description:\n- resolution threshold: 5 angstrom,\n- PDB snapshot: 20190101,\n- structure methods accepted: all (x-ray christolography, NRM, Cryo-EM),\n- sequence identity threshold for clustering: 40% sequence similarity,\n- maximum length per sequence: 1000 residues,\n- minimum length per sequence: 5 residues,\n- maximum fraction of missing values at the ends: 10%,\n- size of validation subset: 10%.\n\n```bash\nproteinflow generate --tag new --resolution_thr 5 --pdb_snapshot 20190101 --not_filter_methods --min_seq_id 0.4 --max_length 1000 --min_length 5 --missing_ends_thr 0.1 --valid_split 0.1\n```\nSee the [docs](https://adaptyvbio.github.io/ProteinFlow/) (or `proteinflow generate --help`) for the full list of parameters and more information.\n\nA registry of all the files that are removed during the filtering as well as description with the reason for their removal is created automatically for each `generate` command. The log files are save (at `data/logs` by default) and a summary can be accessed running `proteinflow get_summary {log_path}`.\n\n### Running the pipeline (SAbDab)\nYou can also use the `--sabdab` option in `proteinflow generate` to load files from SAbDab and cluster them based on CDRs. By default the `--sabdab` tag will download the latest up-to-date version of the SabDab dataset and cluster the antibodies based on their CDR sequence.\nAlternatively, it can be used together with the tag `--sabdab_data_path` to process a custom SAbDab-like zip file or folder. This allows you to use search and query tools from the [SabDab web interface](https://opig.stats.ox.ac.uk/webapps/newsabdab/sabdab/) to create a custom dataset by downloading the archived zip file of the structures selected. (Under Downloads section of your SabDab query).\n\nSAbDab sequences clustering is done across all 6 Complementary Determining Regions (CDRs) - H1, H2, H3, L1, L2, L3, based on the [Chothia numbering](https://pubmed.ncbi.nlm.nih.gov/9367782/) implemented by SabDab. CDRs from nanobodies and other synthetic constructs are clustered together with other heavy chain CDRs. The resulting CDR clusters are split into training, test and validation in a way that ensures that every PDB file only appears in one subset.\n\nIndividual output pickle files represent heavy chain - light chain - antigen complexes (created from SAbDab annotation, sometimes more than one per PDB entry). Each of the elements (heavy chain, light chain, antigen) can be missing in specific entries and there can be multiple antigen chains. In order to filter for at least one antigen chain, use the `--require_antigen` option.\n\nFor instance, let's generate a dataset with the following description:\n- SabDab version: latest (up-to-date),\n- resolution threshold: 5 angstrom,\n- structure methods accepted: all (x-ray christolography, NRM, Cryo-EM),\n- sequence identity threshold for clustering (CDRs): 40%,\n- size of validation subset: 10%.\n\n```bash\nproteinflow generate --sabdab --tag new --resolution_thr 5 --not_filter_methods --min_seq_id 0.4 --valid_split 0.1\n```\n\n### Splitting\nBy default, both `proteinflow generate` and `proteinflow download` will also split your data into training, test and validation according to MMseqs2 clustering and homomer/heteromer/single chain proportions. However, you can skip this step with a `--skip_splitting` flag and then perform it separately with the `proteinflow split` command.\n\nThe following command will perform the splitting with a 10% validation set, a 5% test set and a 50% threshold for sequence identity clusters.\n```bash\nproteinflow split --tag new --valid_split 0.1 --test_split 0.5 --min_seq_id 0.5\n```\n\nUse the `--exclude_chains` and `--exclude_threshold` parameters to move all biounits that contain chains similar to what you specify to a separate folder.\n\n### Using the data\nThe output files are pickled nested dictionaries where first-level keys are chain Ids and second-level keys are the following:\n- `'crd_bb'`: a `numpy` array of shape `(L, 4, 3)` with backbone atom coordinates (N, C, CA, O),\n- `'crd_sc'`: a `numpy` array of shape `(L, 10, 3)` with sidechain atom coordinates (check `proteinflow.sidechain_order()` for the order of atoms),\n- `'msk'`: a `numpy` array of shape `(L,)` where ones correspond to residues with known coordinates and\n zeros to missing values,\n- `'seq'`: a string of length `L` with residue types.\n\nIn a SAbDab datasets, an additional key is added to the dictionary:\n- `'cdr'`: a `numpy` array of shape `(L,)` where CDR residues are marked with the corresponding type (`'H1'`, `'L1'`, ...) \n and non-CDR residues are marked with `'-'`.\n \nNote that the sequence information in the PDB files is aligned to the FASTA sequences to identify the missing residues.\n\nOnce your data is ready, you can open the files with `pickle` directly.\n\n```python\nimport pickle\nimport os\n\ntrain_folder = \"./data/proteinflow_new/training\"\nfor filename in os.listdir(train_folder):\n with open(os.path.join(train_folder, filename), \"rb\") as f:\n data = pickle.load(f)\n crd_bb = data[\"crd_bb\"]\n seq = data[\"seq\"]\n ...\n```\n\nAlternatively, you can use our `ProteinDataset` or `ProteinLoader` classes \nfor convenient processing. Among other things, they allow for feature extraction, single chain / homomer / heteromer filtering and randomized sampling from sequence identity clusters.\n\nFor example, here is how we can create a data loader that:\n- samples a different cluster representative at every epoch,\n- extracts dihedral angles, sidechain orientation and secondary structure features,\n- only loads pairs of interacting proteins (larger biounits are broken up into pairs),\n- has batch size 8.\n\n```python\nfrom proteinflow import ProteinLoader\ntrain_loader = ProteinLoader.from_args(\n \"./data/proteinflow_new/training\", \n clustering_dict_path=\"./data/proteinflow_new/splits_dict/train.pickle\",\n node_features_type=\"dihedral+sidechain_orientation+secondary_structure\",\n entry_type=\"pair\",\n batch_size=8,\n)\nfor batch in train_loader:\n crd_bb = batch[\"X\"] #(B, L, 4, 3)\n seq = batch[\"S\"] #(B, L)\n sse = batch[\"secondary_structure\"] #(B, L, 3)\n to_predict = batch[\"masked_res\"] #(B, L), 1 where the residues should be masked, 0 otherwise\n ...\n```\nSee more details on available parameters and the data format in the [docs](https://adaptyvbio.github.io/ProteinFlow/) + [this repository](https://github.com/adaptyvbio/ProteinFlow-models) for a use case.\n\n## ProteinFlow Stable Releases\nYou can download them with `proteinflow download --tag {tag}` in the command line or browse in the [interface](https://proteinflow-datasets.s3.eu-west-1.amazonaws.com/index.html).\n\n|Tag |Date |Snapshot|Size|Min res|Min len|Max len|Max chains|MMseqs thr|Split (train/val/test)|Missing thr (ends/middle)|Source|Remove redundancies|Note|\n|-------|--------|--------|----|-------|-------|-------|----------|----------|----------------------|-------------------------|---|---|----------------|\n|paper|10.11.22|20220103|24G|3.5|30|10'000|-|0.3|90/5/5|0.3/0.1|PDB|yes|first release, no mmCIF files|\n|20230102_stable|27.02.23|20230102|28G|3.5|30|10'000|-|0.3|90/5/5|0.3/0.1|PDB|yes|v1.1.1|\n|20230623_sabdab|26.06.23|live 26.06.23|1.4G|3.5|30|10'000|-|0.3|96/3/1|0.5/0.2|SAbDab|no|v1.4.1 (requires >= v1.4.0)|\n|20230102_v200|19.07.23|20230102|33G|3.5|30|10'000|10|0.3|90/5/5|0.3/0.1|PDB|no|v2.0.0|\n|20231221_sabdab|21.12.23|live 21.12.23|1.8G|3.5|30|10'000|-|0.3|96/3/1|0.5/0.2|SAbDab|no|v2.6.1 (requires >= v1.4.0)|\n\n## License\nThe `proteinflow` package and data are released and distributed under the BSD 3-Clause License\n\n\n## Contributions\nThis is an open source project supported by [Adaptyv Bio](https://www.adaptyvbio.com/). Contributions, suggestions and bug-fixes are welcomed.\n\n",
"bugtrack_url": null,
"license": "BSD-3-Clause",
"summary": "Versatile pipeline for processing protein structure data for deep learning applications.",
"version": "2.8.0",
"project_urls": null,
"split_keywords": [
"bioinformatics",
"dataset",
"protein",
"pdb",
"deep learning",
"antibody"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "3d803467f910adce85e16c4dc0131bb13888d791780b3474c507b40af6b25892",
"md5": "26d622f26d3e7037f2e7958c67b88339",
"sha256": "ad3a96691634d01491bccb56a118394ef04be3b4f595349db28730ab5e02db16"
},
"downloads": -1,
"filename": "proteinflow-2.8.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "26d622f26d3e7037f2e7958c67b88339",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 105178,
"upload_time": "2024-02-08T09:49:53",
"upload_time_iso_8601": "2024-02-08T09:49:53.220592Z",
"url": "https://files.pythonhosted.org/packages/3d/80/3467f910adce85e16c4dc0131bb13888d791780b3474c507b40af6b25892/proteinflow-2.8.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "9272aab58ae3836bf1fd3abcf7788ba51a1b45e4d5e5d4640d6b0a69942a04d9",
"md5": "2b7641b1962d2146400f8204f1d4621e",
"sha256": "70d27d6a59c9d35a2daa1f1eaafb6674d532ad367d9ad0def5f8fd0ed277f5c1"
},
"downloads": -1,
"filename": "proteinflow-2.8.0.tar.gz",
"has_sig": false,
"md5_digest": "2b7641b1962d2146400f8204f1d4621e",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 96853,
"upload_time": "2024-02-08T09:49:55",
"upload_time_iso_8601": "2024-02-08T09:49:55.571236Z",
"url": "https://files.pythonhosted.org/packages/92/72/aab58ae3836bf1fd3abcf7788ba51a1b45e4d5e5d4640d6b0a69942a04d9/proteinflow-2.8.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-02-08 09:49:55",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "proteinflow"
}