# SpatialCorr: Analyzing spatially varying correlation


## About
SpatialCorr is a set of statistical methods for identifying genes whose correlation structure changes across a spatial transcriptomics sample. Along with a set of statistical tests, SpatialCorr also offers a number of methods for visualizing spatially varying correlation.
Here is a schematic overview of the analyses performed by SpatialCorr:
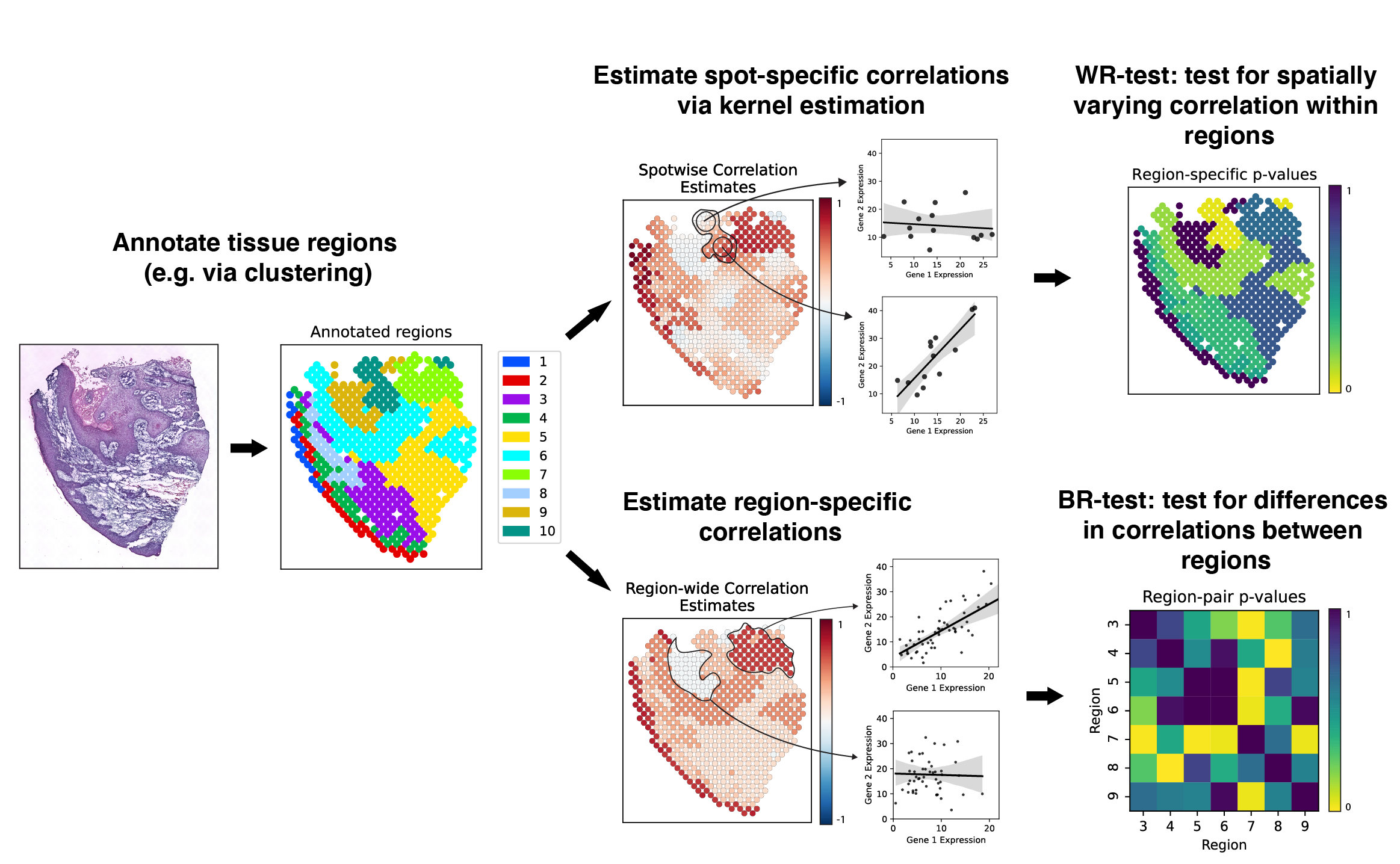
For more details regarding the underlying method, see the paper:
[Bernstein, M.N., Ni, Z., Prasad, A., Brown, J., Mohanty, C., Stewart, R., Newton, M.A., Kendziorski, C. (2022). SpatialCorr: Identifying gene sets with spatially varying correlation structure. *Cell Reports Methods*.](https://doi.org/10.1016/j.crmeth.2022.100369)
## Installation
To install SpatialCorr using Pip, run the following command:
`pip install spatialcorr`
## Usage
#### Documentation
For SpatialCorr's API manual, please visit the [documentation](https://spatialcorr.readthedocs.io/en/latest/index.html).
#### Tutorial
For a tutorial on running SpatialCorr, please see the [tutorial](https://github.com/mbernste/spatialcorr/blob/main/tutorial/SpatialCorr_tutorial.ipynb). This tutorial can also be run via [Google Colab](https://colab.research.google.com/drive/199gpNyyM6Jj8k9LLn1d71l_pX16yko60?usp=sharing).
Raw data
{
"_id": null,
"home_page": "https://github.com/mbernste/spatialcorr",
"name": "spatialcorr",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "spatial-transcriptomics,gene-expression,computational-biology",
"author": "Matthew N. Bernstein",
"author_email": "mbernstein@morgridge.org",
"download_url": "https://files.pythonhosted.org/packages/a0/43/546991112da73440cd6f2a46d3e74a8f876de1a5d693806598ef5abfd1ae/spatialcorr-1.2.0.tar.gz",
"platform": null,
"description": "# SpatialCorr: Analyzing spatially varying correlation \n \n\n\n\n## About\n\nSpatialCorr is a set of statistical methods for identifying genes whose correlation structure changes across a spatial transcriptomics sample. Along with a set of statistical tests, SpatialCorr also offers a number of methods for visualizing spatially varying correlation.\n\nHere is a schematic overview of the analyses performed by SpatialCorr:\n\n\n\nFor more details regarding the underlying method, see the paper: \n[Bernstein, M.N., Ni, Z., Prasad, A., Brown, J., Mohanty, C., Stewart, R., Newton, M.A., Kendziorski, C. (2022). SpatialCorr: Identifying gene sets with spatially varying correlation structure. *Cell Reports Methods*.](https://doi.org/10.1016/j.crmeth.2022.100369)\n\n## Installation\n\nTo install SpatialCorr using Pip, run the following command:\n\n`pip install spatialcorr`\n\n## Usage\n\n#### Documentation\n\nFor SpatialCorr's API manual, please visit the [documentation](https://spatialcorr.readthedocs.io/en/latest/index.html).\n\n#### Tutorial\n\nFor a tutorial on running SpatialCorr, please see the [tutorial](https://github.com/mbernste/spatialcorr/blob/main/tutorial/SpatialCorr_tutorial.ipynb). This tutorial can also be run via [Google Colab](https://colab.research.google.com/drive/199gpNyyM6Jj8k9LLn1d71l_pX16yko60?usp=sharing).\n",
"bugtrack_url": null,
"license": "MIT License",
"summary": "SpatialCorr",
"version": "1.2.0",
"split_keywords": [
"spatial-transcriptomics",
"gene-expression",
"computational-biology"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "45c6290256feb2dfbaf96f44d4bae91948d710ff71f0b8376212bfa1c34cb9e1",
"md5": "83cb6e025b2bcb140d854b5a42f66fd1",
"sha256": "1174a7f3abe9fdea76f4c242c7abc1707834dab07036e90131503c4140280dbe"
},
"downloads": -1,
"filename": "spatialcorr-1.2.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "83cb6e025b2bcb140d854b5a42f66fd1",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": null,
"size": 12872541,
"upload_time": "2023-01-25T15:28:54",
"upload_time_iso_8601": "2023-01-25T15:28:54.842986Z",
"url": "https://files.pythonhosted.org/packages/45/c6/290256feb2dfbaf96f44d4bae91948d710ff71f0b8376212bfa1c34cb9e1/spatialcorr-1.2.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "a043546991112da73440cd6f2a46d3e74a8f876de1a5d693806598ef5abfd1ae",
"md5": "3714175bc8258d10c9e3c762770a78de",
"sha256": "7eb6f356637e8cb323b39dbde5e9fa9f88f790dee02b4d08f68307cfe851101e"
},
"downloads": -1,
"filename": "spatialcorr-1.2.0.tar.gz",
"has_sig": false,
"md5_digest": "3714175bc8258d10c9e3c762770a78de",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 12867473,
"upload_time": "2023-01-25T15:28:57",
"upload_time_iso_8601": "2023-01-25T15:28:57.960040Z",
"url": "https://files.pythonhosted.org/packages/a0/43/546991112da73440cd6f2a46d3e74a8f876de1a5d693806598ef5abfd1ae/spatialcorr-1.2.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-01-25 15:28:57",
"github": true,
"gitlab": false,
"bitbucket": false,
"github_user": "mbernste",
"github_project": "spatialcorr",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"requirements": [
{
"name": "numpy",
"specs": [
[
">=",
"1.19.5"
]
]
},
{
"name": "pandas",
"specs": [
[
">=",
"1.2.1"
]
]
},
{
"name": "scipy",
"specs": [
[
">=",
"1.4.1"
]
]
},
{
"name": "anndata",
"specs": [
[
">=",
"0.7.5"
]
]
},
{
"name": "scikit-learn",
"specs": [
[
">=",
"0.24.2"
]
]
},
{
"name": "matplotlib",
"specs": []
},
{
"name": "seaborn",
"specs": [
[
">=",
"0.11.1"
]
]
},
{
"name": "statsmodels",
"specs": [
[
">=",
"0.12.2"
]
]
},
{
"name": "numpydoc",
"specs": [
[
">=",
"1.1.0"
]
]
}
],
"lcname": "spatialcorr"
}
