<p align="center">
<img src="https://raw.githubusercontent.com/cgoliver/rnaglib/master/images/rgl.png#gh-light-mode-only" width="30%">
</p>
# RNA Geometric Library (`rnaglib`)
<div align="center">

[](https://pypi.org/project/rnaglib/)
[](https://rnaglib.readthedocs.io/en/latest/?badge=latest)
[](https://codecov.io/gh/cgoliver/rnaglib)
</div>
`RNAglib` is a Python package for studying RNA 2.5D and 3D structures. Functionality includes automated data loading,
analysis, visualization, ML model building and benchmarking.

A web-based documentation is available at [**rnaglib.org**](https://rnaglib.org).
We host RNAs annotated with molecule, base pair, and nucleotide level attributes. These include, but are not limited to:
* Secondary structure and 3D coordinates
* Leontis-Westhof base pair geometry classification
* Protein binding, small molecule binding, chemical modifications...
To install the tool, follow the steps in [INSTALL.md](INSTALL.md).
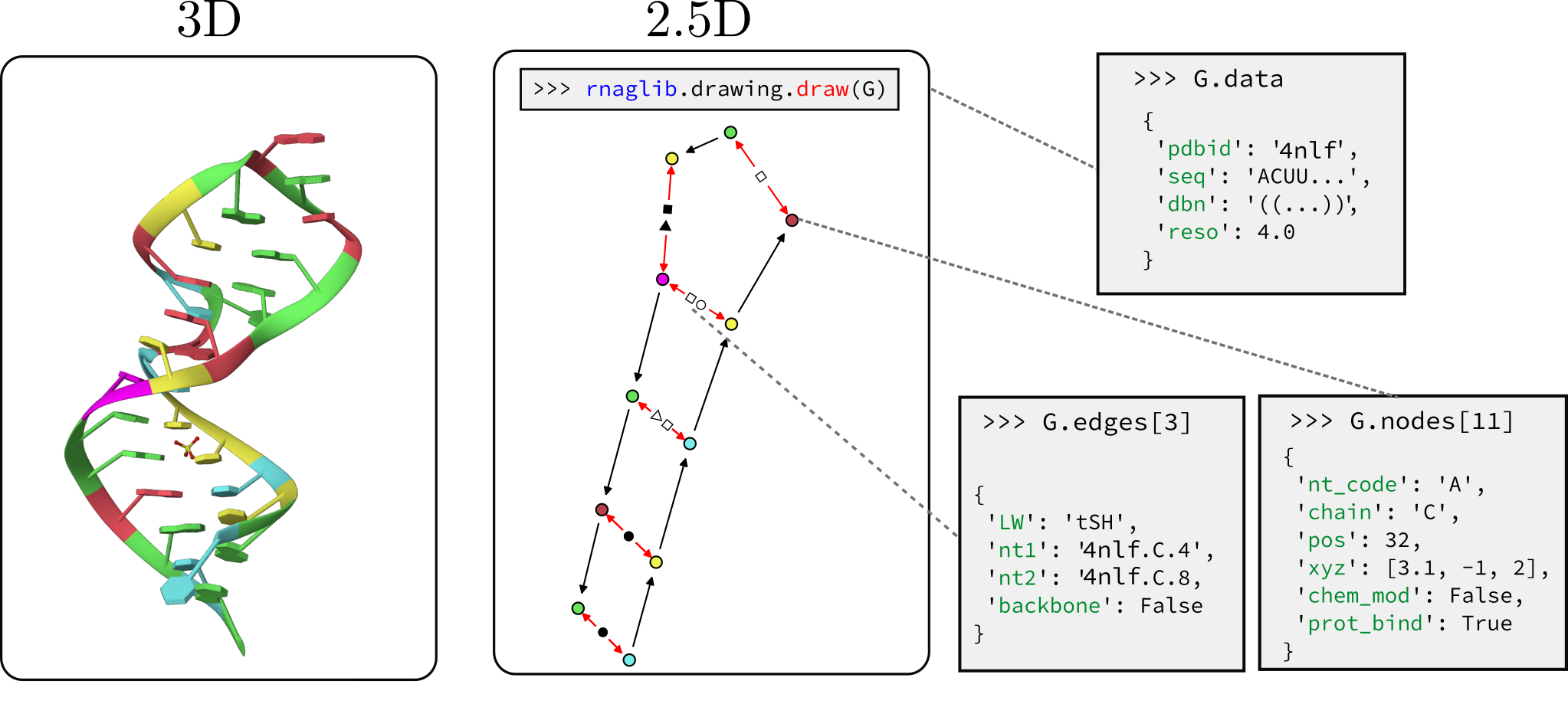
## What can you do with `rnaglib`?
A quickstart and tutorials are available in our online documentation: [**rnaglib.org**](https://rnaglib.org).
In this readme we briefly review the functionality of rnaglib:
- [Benchmark ML models](#benchmark-ml-models-on-rna-3d-structures-new)
- [Get annotated RNA 3D structures](#get-annotated-rna-3d-structures)
- [Fetch and browse annotated RNA 3D structures](#fetch-and-browse-annotated-rna-3D-structures)
- [Dowloading whole RNA structure databases](#Dowloading-whole-RNA-structure-databases)
- [Annotate your own structures](#Annotate-your-own-structures)
- [Additional functionalities](#Additional-functionalities)
- [Quick visualization of 2.5D graphs](#Quick-visualization-of-2.5D-graphs)
- [2.5D graph comparison and alignment](#2.5D-graph-comparison-and-alignment)
- [Citing the tool](#citation)
- [Around RNAglib](#Around-RNAglib)
## Benchmark ML models on RNA 3D structures (**new**)
We now provide datasets of RNA 3D structures ready-to-use for machine learning model benchmarking in seven
biologically relevant tasks.
Moreover, we provide many tools to create your own new tasks.
A more detailed description is provided in the [Tasks' README ](src/rnaglib/tasks/README.md) and in the
[documentation](https://rnaglib.org/en/latest/tutorials/tuto_tasks.html).
Everything you need to train and evaluate a model is built on 3 basic ingredients:
1. A ``rnaglib.Task`` object with holds all the relevant data, splits and functionality.
2. A ``rnaglib.Representation`` object which converts raw RNAs to tensor formats.
3. A model of your choosing, though we provide a basic one to get started ``rnaglib.learning.PyGmodel``
```python
from rnaglib.tasks import ChemicalModification
from rnaglib.transforms import GraphRepresentation
from rnaglib.learning.task_models import PygModel
# Load task, representation, and get loaders
task = ChemicalModification(root="my_root")
model = PygModel.from_task(task)
pyg_rep = GraphRepresentation(framework="pyg")
task.add_representation(pyg_rep)
train_loader, val_loader, test_loader = task.get_split_loaders(batch_size=8)
for batch in train_loader:
batch = batch['graph'].to(model.device)
output = model(batch)
test_metrics = model.evaluate(task, split='test')
```
## Get annotated RNA 3D structures
### Fetch and browse annotated RNA 3D structures
Current release contains annotations generated by x3dna-dssr as well as some additional ones that we added for all
available PDBs at the time of release.
Each RNA is stored as a networkx graph where nodes are residues and edges are backbone and base pairing edges.
The networkx graph object has graph-level, node-level and edge-level attributes.
[Here](https://rnaglib.org/en/latest/rna_ref.html) is a reference for all the annotations currently
available.
```python
>>> from rnaglib.dataset import rna_from_pdbid
>>> rna_dict = rna_from_pdbid('1fmn') # fetch from local database or RCSB if not found
>>> rna_dict['rna'].graph # display graph-level features
{'name': '1fmn', 'pdbid': '1fmn', 'ligand_to_smiles': {'FMN': 'Cc1cc2c(cc1C)N(C3=NC(=O)NC(=O)C3=N2)CC(C(C(COP(=O)(O)O)O)O)O'}, 'ss': {'A': '..(((((......(((....))).....)))))..'}, 'seq': {'A': 'GGCGUGUAGGAUAUGCUUCGGCAGAAGGACACGCC'}}
```
## Dowloading whole RNA structure databases
In addition to analysing RNA data, RNAglib also distributes available parsed RNA structures.
Databases of annotated structures can be downloaded directly from [Zenodo](https://zenodo.org/records/14625192).
| Version | Date | Total RNAs | Total Non-Redundant | Non-redundant version | `rnaglib` commit |
---------|----------|------------|---------------------|-----------------------|------------------|
2.0.2 | 25-02-25 | 8441 | 2921 | 3.375 | ac303c7 |
2.0.0 | 12-01-25 | 8305 | 2877 | 3.369 | 33a9e989 |
1.0.0 | 15-02-23 | 5759 | 1176 | 3.269 | 5446ae2c |
0.0.0 | 20-07-21 | 3739 | 899 | 3.186 | eb25dabd |
They can also be obtained through the provided command line utility, where you can specify the version and redundancy.
```
$ rnaglib_download -r all|nr
```
## Annotate your own structures
You can extract Leontis-Westhof interactions and convert 3D structures to 2.5D graphs.
We wrap a fork of [fr3d-python](https://github.com/cgoliver/fr3d-python) to support this functionality.
```python
from rnaglib.prepare_data import fr3d_to_graph
G = fr3d_to_graph("../data/structures/1fmn.cif")
```
Warning: this method currently does not support non-standard residues. Support coming soon. Up to version 1.0.0 of the
RNA database were created using x3dna-dssr which do contain non-standard residues.
## Additional functionalities
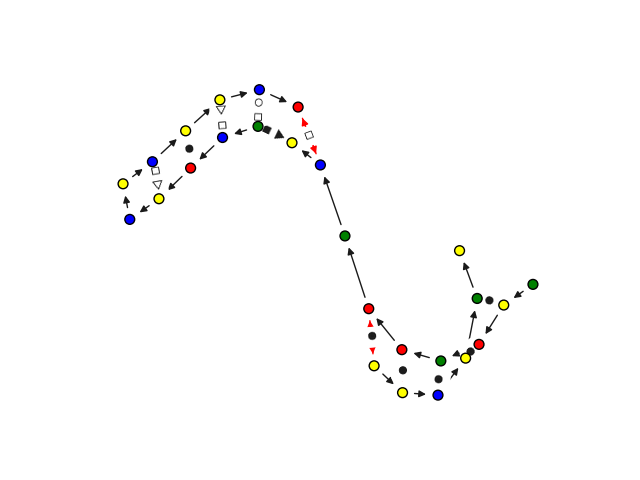
### Quick visualization of 2.5D graphs
We customize networkx graph drawing functionalities to give some convenient visualization of 2.5D base pairing networks.
This is not a dedicated visualization tool, it is only intended for quick debugging. We point you
to [VARNA]()https://varna.lisn.upsaclay.fr/ or [RNAscape](https://academic.oup.com/nar/article/52/W1/W354/7648766) for a
full-featured visualizer.
```python
from rnaglib.drawing import rna_draw
rna_draw(G, show=True, layout="spring")
```
### 2.5D graph comparison and alignment
When dealing with 3D structures as 2.5D graphs we support graph-level comparison through the graph edit distance.
```python
from rnaglib.algorithms import graph_edit_distance
from rnaglib.dataset import rna_from_pdbid
G = rna_from_pdbid("4nlf")["rna"]
print(graph_edit_distance(G, G)) # 0.0
```
## Citation
```
@article{mallet2022rnaglib,
title={RNAglib: a python package for RNA 2.5 D graphs},
author={Mallet, Vincent and Oliver, Carlos and Broadbent, Jonathan and Hamilton, William L and Waldisp{\"u}hl, J{\'e}r{\^o}me},
journal={Bioinformatics},
volume={38},
number={5},
pages={1458--1459},
year={2022},
publisher={Oxford University Press}
}
```
## Around RNAglib
### Projects using `rnaglib`
If you use rnaglib in one of your projects, please cite and feel free to make a pull request so we can list your project
here.
* [RNAMigos2](https://github.com/cgoliver/RNAmigos2)
* [Structure-and Function-Aware Substitution Matrices](https://github.com/BorgwardtLab/GraphMatchingSubstitutionMatrices)
* [MultiModRLBP: A Deep Learning Approach for RNA-Small Molecule Ligand Binding Site Prediction using Multi-modal features](https://github.com/lennylv/MultiModRLBP)
* [VeRNAl](https://github.com/cgoliver/vernal)
* [RNAMigos](https://github.com/cgoliver/RNAmigos)
### Resources
* [Documentation](https://rnaglib.readthedocs.io/en/latest/?badge=latest)
* [Twitter](https://twitter.com/rnaglib)
* Contact: `rnaglib@cs.mcgill.ca`
### References
1. Leontis, N. B., & Zirbel, C. L. (2012). Nonredundant 3D Structure Datasets for RNA Knowledge Extraction and
Benchmarking. In RNA 3D Structure Analysis and Prediction N. Leontis & E. Westhof (Eds.), (Vol. 27, pp. 281–298).
Springer Berlin Heidelberg. doi:10.1007/978-3-642-25740-7\_13
Raw data
{
"_id": null,
"home_page": null,
"name": "rnaglib",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.7",
"maintainer_email": null,
"keywords": "RNA, 3D, graph neural network",
"author": null,
"author_email": "Vincent Mallet <vincentx15@gmail.com>, Carlos Oliver <oliver@biochem.mpg.de>, Jonathan Broadbent <jonathan.broadbent@mail.utoronto.ca>, \"William L. Hamilton\" <wlh@cs.mcgill.ca>, J\u00e9rome Waldispuhl <jeromew@cs.mcgill.ca>",
"download_url": "https://files.pythonhosted.org/packages/6f/77/f25973f93f0ceca0c5dad42a198ab1c004d1e9a73e71c63b69265b2a3883/rnaglib-3.4.7.tar.gz",
"platform": null,
"description": "<p align=\"center\">\n<img src=\"https://raw.githubusercontent.com/cgoliver/rnaglib/master/images/rgl.png#gh-light-mode-only\" width=\"30%\">\n</p>\n\n# RNA Geometric Library (`rnaglib`)\n\n<div align=\"center\">\n\n\n[](https://pypi.org/project/rnaglib/)\n[](https://rnaglib.readthedocs.io/en/latest/?badge=latest)\n[](https://codecov.io/gh/cgoliver/rnaglib)\n</div>\n\n`RNAglib` is a Python package for studying RNA 2.5D and 3D structures. Functionality includes automated data loading,\nanalysis, visualization, ML model building and benchmarking.\n\n\n\nA web-based documentation is available at [**rnaglib.org**](https://rnaglib.org).\n\nWe host RNAs annotated with molecule, base pair, and nucleotide level attributes. These include, but are not limited to:\n\n* Secondary structure and 3D coordinates\n* Leontis-Westhof base pair geometry classification\n* Protein binding, small molecule binding, chemical modifications...\n\nTo install the tool, follow the steps in [INSTALL.md](INSTALL.md).\n\n\n\n## What can you do with `rnaglib`?\n\nA quickstart and tutorials are available in our online documentation: [**rnaglib.org**](https://rnaglib.org).\nIn this readme we briefly review the functionality of rnaglib:\n\n- [Benchmark ML models](#benchmark-ml-models-on-rna-3d-structures-new)\n- [Get annotated RNA 3D structures](#get-annotated-rna-3d-structures)\n - [Fetch and browse annotated RNA 3D structures](#fetch-and-browse-annotated-rna-3D-structures)\n - [Dowloading whole RNA structure databases](#Dowloading-whole-RNA-structure-databases)\n - [Annotate your own structures](#Annotate-your-own-structures)\n- [Additional functionalities](#Additional-functionalities)\n - [Quick visualization of 2.5D graphs](#Quick-visualization-of-2.5D-graphs)\n - [2.5D graph comparison and alignment](#2.5D-graph-comparison-and-alignment)\n- [Citing the tool](#citation)\n- [Around RNAglib](#Around-RNAglib)\n\n## Benchmark ML models on RNA 3D structures (**new**)\n\nWe now provide datasets of RNA 3D structures ready-to-use for machine learning model benchmarking in seven\nbiologically relevant tasks.\nMoreover, we provide many tools to create your own new tasks.\nA more detailed description is provided in the [Tasks' README ](src/rnaglib/tasks/README.md) and in the\n[documentation](https://rnaglib.org/en/latest/tutorials/tuto_tasks.html). \n\nEverything you need to train and evaluate a model is built on 3 basic ingredients:\n\n1. A ``rnaglib.Task`` object with holds all the relevant data, splits and functionality.\n2. A ``rnaglib.Representation`` object which converts raw RNAs to tensor formats.\n3. A model of your choosing, though we provide a basic one to get started ``rnaglib.learning.PyGmodel``\n\n```python\nfrom rnaglib.tasks import ChemicalModification\nfrom rnaglib.transforms import GraphRepresentation\nfrom rnaglib.learning.task_models import PygModel\n\n# Load task, representation, and get loaders\ntask = ChemicalModification(root=\"my_root\")\nmodel = PygModel.from_task(task)\npyg_rep = GraphRepresentation(framework=\"pyg\")\n\ntask.add_representation(pyg_rep)\ntrain_loader, val_loader, test_loader = task.get_split_loaders(batch_size=8)\n\nfor batch in train_loader:\n batch = batch['graph'].to(model.device)\n output = model(batch)\n\ntest_metrics = model.evaluate(task, split='test')\n```\n\n## Get annotated RNA 3D structures\n\n### Fetch and browse annotated RNA 3D structures\n\nCurrent release contains annotations generated by x3dna-dssr as well as some additional ones that we added for all\navailable PDBs at the time of release.\n\nEach RNA is stored as a networkx graph where nodes are residues and edges are backbone and base pairing edges.\nThe networkx graph object has graph-level, node-level and edge-level attributes.\n[Here](https://rnaglib.org/en/latest/rna_ref.html) is a reference for all the annotations currently\navailable.\n\n```python\n\n>>> from rnaglib.dataset import rna_from_pdbid\n>>> rna_dict = rna_from_pdbid('1fmn') # fetch from local database or RCSB if not found\n>>> rna_dict['rna'].graph # display graph-level features\n{'name': '1fmn', 'pdbid': '1fmn', 'ligand_to_smiles': {'FMN': 'Cc1cc2c(cc1C)N(C3=NC(=O)NC(=O)C3=N2)CC(C(C(COP(=O)(O)O)O)O)O'}, 'ss': {'A': '..(((((......(((....))).....)))))..'}, 'seq': {'A': 'GGCGUGUAGGAUAUGCUUCGGCAGAAGGACACGCC'}}\n```\n\n## Dowloading whole RNA structure databases\n\nIn addition to analysing RNA data, RNAglib also distributes available parsed RNA structures.\nDatabases of annotated structures can be downloaded directly from [Zenodo](https://zenodo.org/records/14625192).\n\n| Version | Date | Total RNAs | Total Non-Redundant | Non-redundant version | `rnaglib` commit |\n---------|----------|------------|---------------------|-----------------------|------------------|\n 2.0.2 | 25-02-25 | 8441 | 2921 | 3.375 | ac303c7 |\n 2.0.0 | 12-01-25 | 8305 | 2877 | 3.369 | 33a9e989 |\n 1.0.0 | 15-02-23 | 5759 | 1176 | 3.269 | 5446ae2c |\n 0.0.0 | 20-07-21 | 3739 | 899 | 3.186 | eb25dabd |\n\nThey can also be obtained through the provided command line utility, where you can specify the version and redundancy.\n\n```\n$ rnaglib_download -r all|nr\n```\n\n## Annotate your own structures\n\nYou can extract Leontis-Westhof interactions and convert 3D structures to 2.5D graphs.\nWe wrap a fork of [fr3d-python](https://github.com/cgoliver/fr3d-python) to support this functionality.\n\n```python\nfrom rnaglib.prepare_data import fr3d_to_graph\n\nG = fr3d_to_graph(\"../data/structures/1fmn.cif\")\n```\n\nWarning: this method currently does not support non-standard residues. Support coming soon. Up to version 1.0.0 of the\nRNA database were created using x3dna-dssr which do contain non-standard residues.\n\n## Additional functionalities\n\n### Quick visualization of 2.5D graphs\n\nWe customize networkx graph drawing functionalities to give some convenient visualization of 2.5D base pairing networks.\nThis is not a dedicated visualization tool, it is only intended for quick debugging. We point you\nto [VARNA]()https://varna.lisn.upsaclay.fr/ or [RNAscape](https://academic.oup.com/nar/article/52/W1/W354/7648766) for a\nfull-featured visualizer.\n\n```python\nfrom rnaglib.drawing import rna_draw\n\nrna_draw(G, show=True, layout=\"spring\")\n```\n\n\n\n### 2.5D graph comparison and alignment\n\nWhen dealing with 3D structures as 2.5D graphs we support graph-level comparison through the graph edit distance.\n\n```python\nfrom rnaglib.algorithms import graph_edit_distance\nfrom rnaglib.dataset import rna_from_pdbid\n\nG = rna_from_pdbid(\"4nlf\")[\"rna\"]\nprint(graph_edit_distance(G, G)) # 0.0\n```\n\n## Citation\n\n```\n@article{mallet2022rnaglib,\n title={RNAglib: a python package for RNA 2.5 D graphs},\n author={Mallet, Vincent and Oliver, Carlos and Broadbent, Jonathan and Hamilton, William L and Waldisp{\\\"u}hl, J{\\'e}r{\\^o}me},\n journal={Bioinformatics},\n volume={38},\n number={5},\n pages={1458--1459},\n year={2022},\n publisher={Oxford University Press}\n}\n```\n\n## Around RNAglib\n\n### Projects using `rnaglib`\n\nIf you use rnaglib in one of your projects, please cite and feel free to make a pull request so we can list your project\nhere.\n\n* [RNAMigos2](https://github.com/cgoliver/RNAmigos2)\n* [Structure-and Function-Aware Substitution Matrices](https://github.com/BorgwardtLab/GraphMatchingSubstitutionMatrices)\n* [MultiModRLBP: A Deep Learning Approach for RNA-Small Molecule Ligand Binding Site Prediction using Multi-modal features](https://github.com/lennylv/MultiModRLBP)\n* [VeRNAl](https://github.com/cgoliver/vernal)\n* [RNAMigos](https://github.com/cgoliver/RNAmigos)\n\n### Resources\n\n* [Documentation](https://rnaglib.readthedocs.io/en/latest/?badge=latest)\n* [Twitter](https://twitter.com/rnaglib)\n* Contact: `rnaglib@cs.mcgill.ca`\n\n### References\n\n1. Leontis, N. B., & Zirbel, C. L. (2012). Nonredundant 3D Structure Datasets for RNA Knowledge Extraction and\n Benchmarking. In RNA 3D Structure Analysis and Prediction N. Leontis & E. Westhof (Eds.), (Vol. 27, pp. 281\u2013298).\n Springer Berlin Heidelberg. doi:10.1007/978-3-642-25740-7\\_13\n\n",
"bugtrack_url": null,
"license": "MIT License",
"summary": "RNAglib: Tools for learning on the structure of RNA using 2.5D geometric representations",
"version": "3.4.7",
"project_urls": {
"Documentation": "https://rnaglib.readthedocs.io/en/latest/index.html",
"GitHub": "https://github.com/cgoliver/rnaglib"
},
"split_keywords": [
"rna",
" 3d",
" graph neural network"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "43646e21a579637a771e4f192c5cccd67851bb1b382654e5fb321e023169a33e",
"md5": "5d1f69ca2ec26e0982245793f528ecc2",
"sha256": "bcec901299abc8daabe9834abf9eb01d69d70a4e9f5ac4931cf392d5e15a813c"
},
"downloads": -1,
"filename": "rnaglib-3.4.7-py3-none-any.whl",
"has_sig": false,
"md5_digest": "5d1f69ca2ec26e0982245793f528ecc2",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.7",
"size": 3115666,
"upload_time": "2025-07-08T21:15:28",
"upload_time_iso_8601": "2025-07-08T21:15:28.492396Z",
"url": "https://files.pythonhosted.org/packages/43/64/6e21a579637a771e4f192c5cccd67851bb1b382654e5fb321e023169a33e/rnaglib-3.4.7-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "6f77f25973f93f0ceca0c5dad42a198ab1c004d1e9a73e71c63b69265b2a3883",
"md5": "8d5f871b7beac7315fd1da53815de73e",
"sha256": "a1a09923ac72bbab9fec983c982227407b00a964c79e6366ebde964a9b9081c0"
},
"downloads": -1,
"filename": "rnaglib-3.4.7.tar.gz",
"has_sig": false,
"md5_digest": "8d5f871b7beac7315fd1da53815de73e",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.7",
"size": 2728571,
"upload_time": "2025-07-08T21:15:30",
"upload_time_iso_8601": "2025-07-08T21:15:30.492820Z",
"url": "https://files.pythonhosted.org/packages/6f/77/f25973f93f0ceca0c5dad42a198ab1c004d1e9a73e71c63b69265b2a3883/rnaglib-3.4.7.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-07-08 21:15:30",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "cgoliver",
"github_project": "rnaglib",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [
{
"name": "biopython",
"specs": []
},
{
"name": "bidict",
"specs": []
},
{
"name": "cython",
"specs": []
},
{
"name": "forgi",
"specs": []
},
{
"name": "fr3d",
"specs": []
},
{
"name": "gemmi",
"specs": []
},
{
"name": "joblib",
"specs": []
},
{
"name": "loguru",
"specs": []
},
{
"name": "networkx",
"specs": []
},
{
"name": "numpy",
"specs": []
},
{
"name": "PuLP",
"specs": []
},
{
"name": "pytest",
"specs": []
},
{
"name": "requests",
"specs": []
},
{
"name": "rna-fm",
"specs": []
},
{
"name": "rdkit",
"specs": []
},
{
"name": "seaborn",
"specs": []
},
{
"name": "scikit-learn",
"specs": []
},
{
"name": "torch",
"specs": []
},
{
"name": "torch_geometric",
"specs": []
},
{
"name": "tqdm",
"specs": []
}
],
"lcname": "rnaglib"
}
