# SpatialCell: Integrated Spatial Transcriptomics Analysis Pipeline
[](https://github.com/Xinyan-C/Spatialcell)
[](https://www.python.org/downloads/)
[](https://opensource.org/licenses/Apache-2.0)
[](https://github.com/Xinyan-C/Spatialcell/issues)
**SpatialCell** is an integrated computational pipeline for spatial transcriptomics analysis that combines cell segmentation and automated cell type annotation. It seamlessly integrates **Stardist (applied as QuPath plugin for cell detection)** for histological image analysis, **Bin2cell** for spatial cell segmentation, and **TopAct** for machine learning-based cell classification.
## 🚀 Key Features
- **Multi-scale Cell Segmentation**: Stardist-enabled QuPath cell detection with Bin2cell spatial segmentation
- **Automated Cell Annotation**: TopAct-based machine learning classification
- **ROI-aware Processing**: Region-of-interest focused analysis for large datasets
- **Scalable Pipeline**: Support for multiple developmental time points (e.g., E14.5, E18.5, P3) and samples
- **Visualization Tools**: Comprehensive plotting and export capabilities
- **Modular Design**: Easy to customize and extend for specific research needs
## 🔧 Installation
### Prerequisites
- Python 3.10 or higher
- QuPath (for histological image analysis)
- Git
- Operating Systems tested: Ubuntu 22.04.03, MacOS 15.5
- Hardware: Standard desktop CPU; GPU not required but optional for accelerated image processing
- Additional Python dependencies are listed in `requirements.txt`
### Typical installation time
Installation usually completes within 5 minutes on a stable internet connection and a typical desktop computer.
### Quick Install (Recommended)
To enable full functionality including TopAct classification, please install TopAct separately:
```bash
pip install spatialcell
pip install git+https://gitlab.com/kfbenjamin/topact.git
```
### Alternative: Install from Source
```bash
# Clone the repository
git clone https://github.com/Xinyan-C/Spatialcell.git
cd Spatialcell
# Install dependencies
pip install -r requirements.txt
# Install the package in editable mode
pip install -e .
```
## 📋 Demo Data and tutorial notebook
The `examples/` directory contains the tutorial notebook to quickly test and understand SpatialCell.
Demo datasets for E14.5, E18.5, and P3 are archived on Zenodo (https://zenodo.org/records/16400171)
### Expected output
- **ROI coordinates** saved as a `.txt` file
- e.g. `examples/demo_data/E18.5_ranges.txt`
- **Binary segmentation masks** saved as `.npz` files
- e.g. `examples/demo_data/E18.5_qupath.npz`
- **Spatial segmentation results** under `examples/demo_data/demo_output/`(more information at https://github.com/Teichlab/bin2cell.git):
- **Data/**
- `E18.5_2um.h5ad` — AnnData containing 2 μm‐bin counts and coordinates for the entire sample
- `E18.5_b2c.h5ad` — Bin2cell‐reconstructed cell‐level AnnData for the entire sample
- **ROI_Data/** (one subfolder per ROI: CS1, CS2, WT1)
- `{ROI}_adata.h5ad` — Spot‐level AnnData extracted for that specific region
- `{ROI}_cdata.h5ad` — Cell‐level AnnData (Bin2cell output) for that region
- **destripe/**, **expanded_labels/**, **gex_labels/**, **joint_labels/**, **joint_labels_all/**, **npz_labels/**, **render_gex/**, **render_labels/**, **segmentation/**
- PDF reports (quality‐control and visualization overlays) for each processing step
- **Log file**
- `spatial_processing.log` — Records parameters (e.g. `prob_thresh`, `nms_thresh`), runtime info, and warnings
- **Cell annotation outputs** under `examples/demo_data/demo_output/cell_annotation/`:
- `outfile_<sample>_<sample>_-_<ROI>.npy`
- NumPy arrays of per-cell feature matrices (e.g. classification probabilities or aggregated counts) for each ROI (CS1, CS2, WT1)
- `sd_<sample>_<sample>_-_<ROI>.joblib`
- Serialized TopACT classifier models saved after training or calibration on each ROI
- `spatial_data_<sample>_roi.joblib`
- Serialized AnnData object containing spatially indexed spot‐level and cell‐level data passed into TopACT for classification
- **Visualization outputs** under `examples/demo_data/demo_output/visualizations/`:
For each ROI (CS1, CS2, WT1):
- `Spatial_Classification_<sample>_-_<ROI>_overlay.pdf`
- Cell type predictions overlaid directly on the high‐resolution tissue image
- `Spatial_Classification_<sample>_-_<ROI>_side_by_side.pdf`
- Side-by-side panels showing (left) raw segmentation mask and (right) classification overlay for comparison
- `Spatial_Classification_<sample>_-_<ROI>.pdf`
- High-resolution, publication-ready map of predicted cell types (colored segmentation)
### Runtime estimate
Approximately 30-45 minutes on a standard desktop for the demo dataset.
## 📖 Usage Instructions
The easiest way to accomplish the pipeline is with our Jupyter notebook tutorial, the tutorial covers the complete workflow from ROI extraction to visualization.
## 🗂️ Project Structure
```
Spatialcell/
├── spatialcell/ # Main package
│ ├── qupath_scripts/ # QuPath-Stardist integration scripts
│ ├── preprocessing/ # Data preprocessing modules
│ ├── spatial_segmentation/ # Bin2cell integration
│ ├── cell_annotation/ # TopAct classification
│ └── utils/ # Utility functions
├── examples/ # Tutorial notebook
│ └── SpatialCell_Demo.ipynb # Jupyter notebooks for tutorial and article reproducibility
├── requirements.txt # Python dependencies
├── setup.py # Package installation script
└── README.md # This file
```
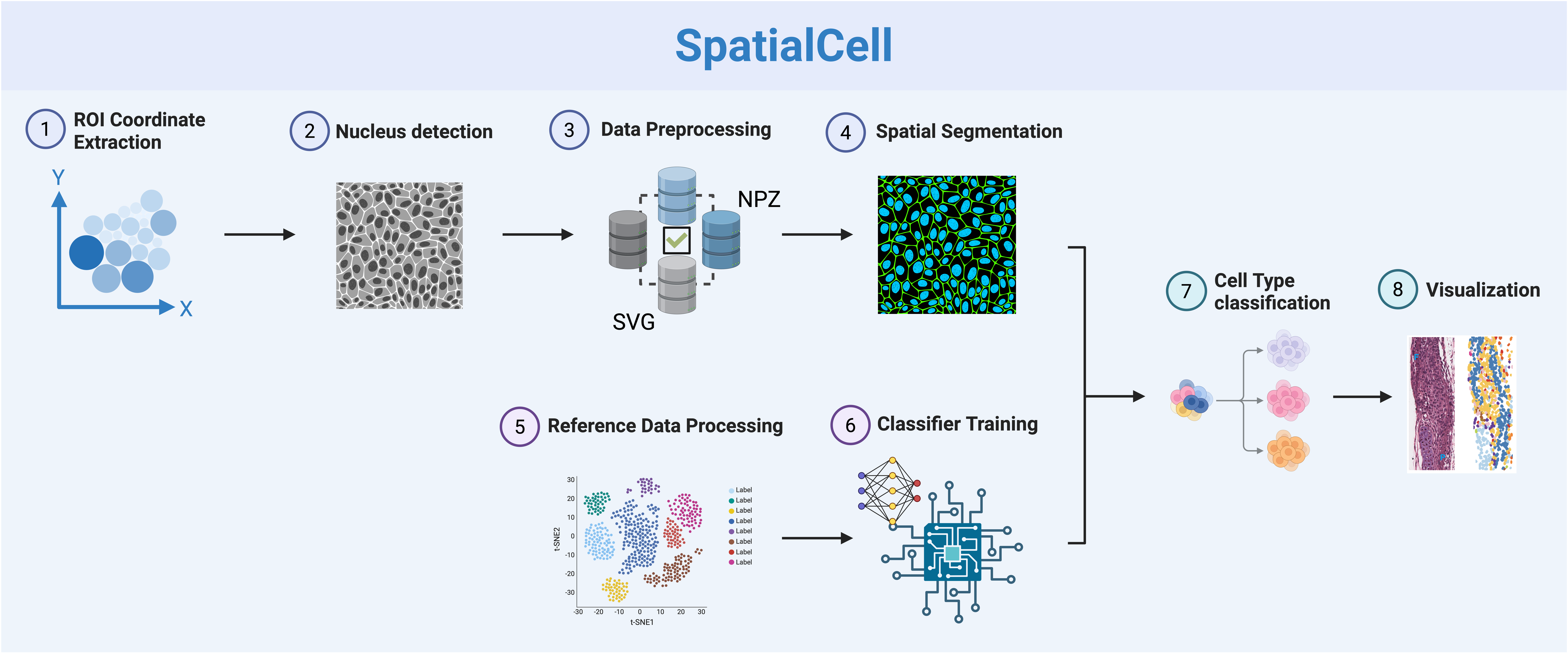
## 🔬 Workflow Overview
1. **ROI Coordinate Extraction**: Extract region-of-interest coordinates from Loupe Browser exports
2. **Nucleus detection**: StarDist-based nucleus detection via QuPath with SVG export
3. **Data Preprocessing**: SVG to NPZ conversion and label mask generation
4. **Spatial Segmentation**: Bin2cell integration with nucleus boundaries and label expansion
5. **Reference Data Processing**: Extract training data from Seurat RDS files
6. **Classifier Training**: Train TopAct machine learning models for cell type annotation
7. **Cell Type Classification**: Apply TopAct classifiers for spatial cell type prediction
8. **Comprehensive Visualization**: Multi-scale plotting, overlay generation, and result export
## 📝 License
SpatialCell is licensed under the **Apache License 2.0**, which includes patent protection and allows commercial use.
### Dependency Licenses:
- **bin2cell**: MIT License (automatically installed)
- **TopAct**: GPL v3 License (optional, user installs separately)
Note: Users should be aware of GPL license requirements when installing TopAct.
For full license text, see the [LICENSE](https://github.com/Xinyan-C/Spatialcell/blob/main/LICENSE) file.
## 📚 Article reproducibility
Jupyter notebooks (e.g. `examples/SpatialCell_Demo.ipynb`) needed to reproduce our analyses in the article *Spatiotemporal Single-Cell Atlas of Suture Stem Cell Dynamics in Craniosynostosis* are included in the `examples/` directory. A minimal example dataset for E14.5, E18.5, and P3 is archived on Zenodo (https://zenodo.org/records/16400171).
## 📄 Citation
If you use SpatialCell in your research, please cite:
```bibtex
@software{spatialcell2025,
author = {Xinyan},
title = {SpatialCell: Integrated Spatial Transcriptomics Analysis Pipeline},
url = {https://github.com/Xinyan-C/Spatialcell},
year = {2025}
}
```
## 📧 Contact
- **Author**: Xinyan
- **Email**: keepandon@gmail.com
- **GitHub**: [@Xinyan-C](https://github.com/Xinyan-C)
## 🔗 References
- **QuPath**: Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi:10.1038/s41598-017-17204-5
- **Stardist**: Schmidt U, Weigert M, Broaddus C, Myers G. Cell detection with star-convex polygons. MICCAI 2018: 265-273. doi:10.1007/978-3-030-00934-2_30
- **Bin2cell**: Polański K, Bartolomé-Casado R, Sarropoulos I, et al. Bin2cell reconstructs cells from high resolution visium HD data. Bioinformatics. 2024;40(9):btae546. doi:10.1093/bioinformatics/btae546
- **TopAct**: Benjamin K, Bhandari A, Kepple JD, et al. Multiscale topology classifies cells in subcellular spatial transcriptomics. Nature. 2024;630(8018):943-949. doi:10.1038/s41586-024-07563-1
- **Scanpy**: Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19(1):15. doi:10.1186/s13059-017-1382-0
Raw data
{
"_id": null,
"home_page": "https://github.com/Xinyan-C/Spatialcell",
"name": "spatialcell",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": null,
"keywords": "spatial-transcriptomics, cell-segmentation, cell-annotation, QuPath, Bin2cell, TopAct, bioinformatics, single-cell",
"author": "Xinyan",
"author_email": "Xinyan <keepandon@gmail.com>",
"download_url": "https://files.pythonhosted.org/packages/13/e3/4e2f22d5519552df9dd68ebbcc39b75fced3faf650b35f5e3302925fb0d0/spatialcell-1.1.1.tar.gz",
"platform": null,
"description": "# SpatialCell: Integrated Spatial Transcriptomics Analysis Pipeline\n\n[](https://github.com/Xinyan-C/Spatialcell)\n\n[](https://www.python.org/downloads/)\n[](https://opensource.org/licenses/Apache-2.0)\n[](https://github.com/Xinyan-C/Spatialcell/issues)\n\n**SpatialCell** is an integrated computational pipeline for spatial transcriptomics analysis that combines cell segmentation and automated cell type annotation. It seamlessly integrates **Stardist (applied as QuPath plugin for cell detection)** for histological image analysis, **Bin2cell** for spatial cell segmentation, and **TopAct** for machine learning-based cell classification.\n\n## \ud83d\ude80 Key Features\n\n- **Multi-scale Cell Segmentation**: Stardist-enabled QuPath cell detection with Bin2cell spatial segmentation \n- **Automated Cell Annotation**: TopAct-based machine learning classification \n- **ROI-aware Processing**: Region-of-interest focused analysis for large datasets \n- **Scalable Pipeline**: Support for multiple developmental time points (e.g., E14.5, E18.5, P3) and samples \n- **Visualization Tools**: Comprehensive plotting and export capabilities \n- **Modular Design**: Easy to customize and extend for specific research needs \n\n## \ud83d\udd27 Installation\n\n### Prerequisites\n\n- Python 3.10 or higher \n- QuPath (for histological image analysis) \n- Git \n- Operating Systems tested: Ubuntu 22.04.03, MacOS 15.5 \n- Hardware: Standard desktop CPU; GPU not required but optional for accelerated image processing \n- Additional Python dependencies are listed in `requirements.txt`\n\n### Typical installation time\n\nInstallation usually completes within 5 minutes on a stable internet connection and a typical desktop computer.\n\n### Quick Install (Recommended)\nTo enable full functionality including TopAct classification, please install TopAct separately:\n\n```bash\npip install spatialcell\npip install git+https://gitlab.com/kfbenjamin/topact.git\n```\n\n### Alternative: Install from Source\n\n```bash\n# Clone the repository\ngit clone https://github.com/Xinyan-C/Spatialcell.git\ncd Spatialcell\n\n# Install dependencies\npip install -r requirements.txt\n\n# Install the package in editable mode\npip install -e .\n```\n\n## \ud83d\udccb Demo Data and tutorial notebook\n\nThe `examples/` directory contains the tutorial notebook to quickly test and understand SpatialCell. \nDemo datasets for E14.5, E18.5, and P3 are archived on Zenodo (https://zenodo.org/records/16400171)\n\n\n### Expected output\n\n- **ROI coordinates** saved as a `.txt` file \n - e.g. `examples/demo_data/E18.5_ranges.txt` \n\n- **Binary segmentation masks** saved as `.npz` files \n - e.g. `examples/demo_data/E18.5_qupath.npz` \n\n- **Spatial segmentation results** under `examples/demo_data/demo_output/`(more information at https://github.com/Teichlab/bin2cell.git): \n - **Data/** \n - `E18.5_2um.h5ad` \u2014 AnnData containing 2 \u03bcm\u2010bin counts and coordinates for the entire sample \n - `E18.5_b2c.h5ad` \u2014 Bin2cell\u2010reconstructed cell\u2010level AnnData for the entire sample \n - **ROI_Data/** (one subfolder per ROI: CS1, CS2, WT1) \n - `{ROI}_adata.h5ad` \u2014 Spot\u2010level AnnData extracted for that specific region \n - `{ROI}_cdata.h5ad` \u2014 Cell\u2010level AnnData (Bin2cell output) for that region \n - **destripe/**, **expanded_labels/**, **gex_labels/**, **joint_labels/**, **joint_labels_all/**, **npz_labels/**, **render_gex/**, **render_labels/**, **segmentation/** \n - PDF reports (quality\u2010control and visualization overlays) for each processing step \n - **Log file** \n - `spatial_processing.log` \u2014 Records parameters (e.g. `prob_thresh`, `nms_thresh`), runtime info, and warnings \n\n- **Cell annotation outputs** under `examples/demo_data/demo_output/cell_annotation/`: \n - `outfile_<sample>_<sample>_-_<ROI>.npy` \n - NumPy arrays of per-cell feature matrices (e.g. classification probabilities or aggregated counts) for each ROI (CS1, CS2, WT1) \n - `sd_<sample>_<sample>_-_<ROI>.joblib` \n - Serialized TopACT classifier models saved after training or calibration on each ROI \n - `spatial_data_<sample>_roi.joblib` \n - Serialized AnnData object containing spatially indexed spot\u2010level and cell\u2010level data passed into TopACT for classification \n\n- **Visualization outputs** under `examples/demo_data/demo_output/visualizations/`: \n For each ROI (CS1, CS2, WT1): \n - `Spatial_Classification_<sample>_-_<ROI>_overlay.pdf` \n - Cell type predictions overlaid directly on the high\u2010resolution tissue image \n - `Spatial_Classification_<sample>_-_<ROI>_side_by_side.pdf` \n - Side-by-side panels showing (left) raw segmentation mask and (right) classification overlay for comparison \n - `Spatial_Classification_<sample>_-_<ROI>.pdf` \n - High-resolution, publication-ready map of predicted cell types (colored segmentation) \n\n### Runtime estimate\n\nApproximately 30-45 minutes on a standard desktop for the demo dataset.\n\n## \ud83d\udcd6 Usage Instructions\n\nThe easiest way to accomplish the pipeline is with our Jupyter notebook tutorial, the tutorial covers the complete workflow from ROI extraction to visualization.\n\n## \ud83d\uddc2\ufe0f Project Structure\n\n```\nSpatialcell/\n\u251c\u2500\u2500 spatialcell/ # Main package\n\u2502 \u251c\u2500\u2500 qupath_scripts/ # QuPath-Stardist integration scripts\n\u2502 \u251c\u2500\u2500 preprocessing/ # Data preprocessing modules\n\u2502 \u251c\u2500\u2500 spatial_segmentation/ # Bin2cell integration\n\u2502 \u251c\u2500\u2500 cell_annotation/ # TopAct classification\n\u2502 \u2514\u2500\u2500 utils/ # Utility functions\n\u251c\u2500\u2500 examples/ # Tutorial notebook\n\u2502 \u2514\u2500\u2500 SpatialCell_Demo.ipynb # Jupyter notebooks for tutorial and article reproducibility\n\u251c\u2500\u2500 requirements.txt # Python dependencies\n\u251c\u2500\u2500 setup.py # Package installation script\n\u2514\u2500\u2500 README.md # This file\n```\n\n\n## \ud83d\udd2c Workflow Overview\n\n1. **ROI Coordinate Extraction**: Extract region-of-interest coordinates from Loupe Browser exports\n2. **Nucleus detection**: StarDist-based nucleus detection via QuPath with SVG export\n3. **Data Preprocessing**: SVG to NPZ conversion and label mask generation\n4. **Spatial Segmentation**: Bin2cell integration with nucleus boundaries and label expansion\n5. **Reference Data Processing**: Extract training data from Seurat RDS files \n6. **Classifier Training**: Train TopAct machine learning models for cell type annotation\n7. **Cell Type Classification**: Apply TopAct classifiers for spatial cell type prediction\n8. **Comprehensive Visualization**: Multi-scale plotting, overlay generation, and result export\n\n\n## \ud83d\udcdd License\n\nSpatialCell is licensed under the **Apache License 2.0**, which includes patent protection and allows commercial use.\n\n### Dependency Licenses:\n\n- **bin2cell**: MIT License (automatically installed) \n- **TopAct**: GPL v3 License (optional, user installs separately) \n\nNote: Users should be aware of GPL license requirements when installing TopAct.\n\nFor full license text, see the [LICENSE](https://github.com/Xinyan-C/Spatialcell/blob/main/LICENSE) file.\n\n## \ud83d\udcda Article reproducibility\n\nJupyter notebooks (e.g. `examples/SpatialCell_Demo.ipynb`) needed to reproduce our analyses in the article *Spatiotemporal Single-Cell Atlas of Suture Stem Cell Dynamics in Craniosynostosis* are included in the `examples/` directory. A minimal example dataset for E14.5, E18.5, and P3 is archived on Zenodo (https://zenodo.org/records/16400171).\n\n## \ud83d\udcc4 Citation\n\nIf you use SpatialCell in your research, please cite:\n\n```bibtex\n@software{spatialcell2025,\n author = {Xinyan},\n title = {SpatialCell: Integrated Spatial Transcriptomics Analysis Pipeline},\n url = {https://github.com/Xinyan-C/Spatialcell},\n year = {2025}\n}\n```\n\n## \ud83d\udce7 Contact\n\n- **Author**: Xinyan \n- **Email**: keepandon@gmail.com \n- **GitHub**: [@Xinyan-C](https://github.com/Xinyan-C) \n\n## \ud83d\udd17 References\n\n- **QuPath**: Bankhead P, Loughrey MB, Fern\u00e1ndez JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi:10.1038/s41598-017-17204-5 \n- **Stardist**: Schmidt U, Weigert M, Broaddus C, Myers G. Cell detection with star-convex polygons. MICCAI 2018: 265-273. doi:10.1007/978-3-030-00934-2_30 \n- **Bin2cell**: Pola\u0144ski K, Bartolom\u00e9-Casado R, Sarropoulos I, et al. Bin2cell reconstructs cells from high resolution visium HD data. Bioinformatics. 2024;40(9):btae546. doi:10.1093/bioinformatics/btae546 \n- **TopAct**: Benjamin K, Bhandari A, Kepple JD, et al. Multiscale topology classifies cells in subcellular spatial transcriptomics. Nature. 2024;630(8018):943-949. doi:10.1038/s41586-024-07563-1 \n- **Scanpy**: Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19(1):15. doi:10.1186/s13059-017-1382-0 \n",
"bugtrack_url": null,
"license": null,
"summary": "Integrated pipeline for spatial transcriptomics cell segmentation and annotation using QuPath, Bin2cell, and TopAct",
"version": "1.1.1",
"project_urls": {
"Bug Reports": "https://github.com/Xinyan-C/Spatialcell/issues",
"Changelog": "https://github.com/Xinyan-C/Spatialcell/releases",
"Documentation": "https://github.com/Xinyan-C/Spatialcell#readme",
"Homepage": "https://github.com/Xinyan-C/Spatialcell",
"Repository": "https://github.com/Xinyan-C/Spatialcell",
"Source Code": "https://github.com/Xinyan-C/Spatialcell"
},
"split_keywords": [
"spatial-transcriptomics",
" cell-segmentation",
" cell-annotation",
" qupath",
" bin2cell",
" topact",
" bioinformatics",
" single-cell"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "1c7072f41a7310fc3eada9c405386d002fa780d8e7d0046b4ed9a6630e986252",
"md5": "88e80bb74b8582bc2155245a43f2e8a9",
"sha256": "882994973478fa68069544635e5b1f33cb28dbb68e93ccda327182744753299d"
},
"downloads": -1,
"filename": "spatialcell-1.1.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "88e80bb74b8582bc2155245a43f2e8a9",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 76836,
"upload_time": "2025-07-27T14:07:51",
"upload_time_iso_8601": "2025-07-27T14:07:51.460156Z",
"url": "https://files.pythonhosted.org/packages/1c/70/72f41a7310fc3eada9c405386d002fa780d8e7d0046b4ed9a6630e986252/spatialcell-1.1.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "13e34e2f22d5519552df9dd68ebbcc39b75fced3faf650b35f5e3302925fb0d0",
"md5": "07402db6d72935e2f3165668e9a6479b",
"sha256": "2093f599fd956db23b7f3f29879a37f35c03b335e3babf171bba9ec66010b749"
},
"downloads": -1,
"filename": "spatialcell-1.1.1.tar.gz",
"has_sig": false,
"md5_digest": "07402db6d72935e2f3165668e9a6479b",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 74250,
"upload_time": "2025-07-27T14:07:53",
"upload_time_iso_8601": "2025-07-27T14:07:53.302030Z",
"url": "https://files.pythonhosted.org/packages/13/e3/4e2f22d5519552df9dd68ebbcc39b75fced3faf650b35f5e3302925fb0d0/spatialcell-1.1.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-07-27 14:07:53",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "Xinyan-C",
"github_project": "Spatialcell",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"requirements": [
{
"name": "anndata",
"specs": [
[
"==",
"0.11.3"
]
]
},
{
"name": "anyio",
"specs": [
[
"==",
"4.9.0"
]
]
},
{
"name": "argon2-cffi",
"specs": [
[
"==",
"23.1.0"
]
]
},
{
"name": "argon2-cffi-bindings",
"specs": [
[
"==",
"21.2.0"
]
]
},
{
"name": "aria2",
"specs": [
[
"==",
"0.0.1b0"
]
]
},
{
"name": "array_api_compat",
"specs": [
[
"==",
"1.11.1"
]
]
},
{
"name": "arrow",
"specs": [
[
"==",
"1.3.0"
]
]
},
{
"name": "asttokens",
"specs": []
},
{
"name": "async-lru",
"specs": [
[
"==",
"2.0.5"
]
]
},
{
"name": "attrs",
"specs": [
[
"==",
"25.3.0"
]
]
},
{
"name": "babel",
"specs": [
[
"==",
"2.17.0"
]
]
},
{
"name": "beautifulsoup4",
"specs": [
[
"==",
"4.13.3"
]
]
},
{
"name": "bin2cell",
"specs": [
[
"==",
"0.3.3"
]
]
},
{
"name": "bleach",
"specs": [
[
"==",
"6.2.0"
]
]
},
{
"name": "Bottleneck",
"specs": []
},
{
"name": "bypy",
"specs": [
[
"==",
"1.8.9"
]
]
},
{
"name": "certifi",
"specs": [
[
"==",
"2025.1.31"
]
]
},
{
"name": "cffi",
"specs": [
[
"==",
"1.17.1"
]
]
},
{
"name": "charset-normalizer",
"specs": [
[
"==",
"3.4.1"
]
]
},
{
"name": "comm",
"specs": []
},
{
"name": "contourpy",
"specs": [
[
"==",
"1.3.1"
]
]
},
{
"name": "cramjam",
"specs": []
},
{
"name": "csbdeep",
"specs": [
[
"==",
"0.8.1"
]
]
},
{
"name": "cycler",
"specs": [
[
"==",
"0.12.1"
]
]
},
{
"name": "debugpy",
"specs": []
},
{
"name": "decorator",
"specs": []
},
{
"name": "defusedxml",
"specs": [
[
"==",
"0.7.1"
]
]
},
{
"name": "dill",
"specs": [
[
"==",
"0.4.0"
]
]
},
{
"name": "exceptiongroup",
"specs": []
},
{
"name": "executing",
"specs": []
},
{
"name": "fastjsonschema",
"specs": [
[
"==",
"2.21.1"
]
]
},
{
"name": "fastparquet",
"specs": []
},
{
"name": "fonttools",
"specs": [
[
"==",
"4.56.0"
]
]
},
{
"name": "fqdn",
"specs": [
[
"==",
"1.5.1"
]
]
},
{
"name": "fsspec",
"specs": []
},
{
"name": "h11",
"specs": [
[
"==",
"0.14.0"
]
]
},
{
"name": "h5py",
"specs": [
[
"==",
"3.13.0"
]
]
},
{
"name": "httpcore",
"specs": [
[
"==",
"1.0.7"
]
]
},
{
"name": "httpx",
"specs": [
[
"==",
"0.28.1"
]
]
},
{
"name": "idna",
"specs": [
[
"==",
"3.10"
]
]
},
{
"name": "igraph",
"specs": [
[
"==",
"0.11.8"
]
]
},
{
"name": "imageio",
"specs": [
[
"==",
"2.37.0"
]
]
},
{
"name": "ipykernel",
"specs": []
},
{
"name": "ipython",
"specs": []
},
{
"name": "isoduration",
"specs": [
[
"==",
"20.11.0"
]
]
},
{
"name": "jedi",
"specs": []
},
{
"name": "Jinja2",
"specs": [
[
"==",
"3.1.6"
]
]
},
{
"name": "joblib",
"specs": [
[
"==",
"1.4.2"
]
]
},
{
"name": "json5",
"specs": [
[
"==",
"0.10.0"
]
]
},
{
"name": "jsonpointer",
"specs": [
[
"==",
"3.0.0"
]
]
},
{
"name": "jsonschema",
"specs": [
[
"==",
"4.23.0"
]
]
},
{
"name": "jsonschema-specifications",
"specs": [
[
"==",
"2024.10.1"
]
]
},
{
"name": "jupyter-events",
"specs": [
[
"==",
"0.12.0"
]
]
},
{
"name": "jupyter-lsp",
"specs": [
[
"==",
"2.2.5"
]
]
},
{
"name": "jupyter_client",
"specs": []
},
{
"name": "jupyter_core",
"specs": []
},
{
"name": "jupyter_server",
"specs": [
[
"==",
"2.15.0"
]
]
},
{
"name": "jupyter_server_terminals",
"specs": [
[
"==",
"0.5.3"
]
]
},
{
"name": "jupyterlab",
"specs": [
[
"==",
"4.3.6"
]
]
},
{
"name": "jupyterlab_pygments",
"specs": [
[
"==",
"0.3.0"
]
]
},
{
"name": "jupyterlab_server",
"specs": [
[
"==",
"2.27.3"
]
]
},
{
"name": "kiwisolver",
"specs": [
[
"==",
"1.4.8"
]
]
},
{
"name": "legacy-api-wrap",
"specs": [
[
"==",
"1.4.1"
]
]
},
{
"name": "leidenalg",
"specs": [
[
"==",
"0.10.2"
]
]
},
{
"name": "llvmlite",
"specs": [
[
"==",
"0.44.0"
]
]
},
{
"name": "MarkupSafe",
"specs": [
[
"==",
"3.0.2"
]
]
},
{
"name": "matplotlib",
"specs": [
[
"==",
"3.10.1"
]
]
},
{
"name": "matplotlib-inline",
"specs": []
},
{
"name": "mistune",
"specs": [
[
"==",
"3.1.3"
]
]
},
{
"name": "mkl-service",
"specs": [
[
"==",
"2.4.0"
]
]
},
{
"name": "mkl_fft",
"specs": []
},
{
"name": "mkl_random",
"specs": []
},
{
"name": "multiprocess",
"specs": [
[
"==",
"0.70.18"
]
]
},
{
"name": "natsort",
"specs": [
[
"==",
"8.4.0"
]
]
},
{
"name": "nbclient",
"specs": [
[
"==",
"0.10.2"
]
]
},
{
"name": "nbconvert",
"specs": [
[
"==",
"7.16.6"
]
]
},
{
"name": "nbformat",
"specs": [
[
"==",
"5.10.4"
]
]
},
{
"name": "nest-asyncio",
"specs": []
},
{
"name": "networkx",
"specs": [
[
"==",
"3.4.2"
]
]
},
{
"name": "notebook",
"specs": [
[
"==",
"7.3.3"
]
]
},
{
"name": "notebook_shim",
"specs": [
[
"==",
"0.2.4"
]
]
},
{
"name": "numba",
"specs": [
[
"==",
"0.61.0"
]
]
},
{
"name": "numexpr",
"specs": []
},
{
"name": "numpy",
"specs": []
},
{
"name": "opencv-python",
"specs": [
[
"==",
"4.11.0.86"
]
]
},
{
"name": "overrides",
"specs": [
[
"==",
"7.7.0"
]
]
},
{
"name": "packaging",
"specs": []
},
{
"name": "pandas",
"specs": [
[
"==",
"1.5.3"
]
]
},
{
"name": "pandocfilters",
"specs": [
[
"==",
"1.5.1"
]
]
},
{
"name": "parso",
"specs": []
},
{
"name": "patsy",
"specs": [
[
"==",
"1.0.1"
]
]
},
{
"name": "pexpect",
"specs": []
},
{
"name": "pillow",
"specs": [
[
"==",
"11.1.0"
]
]
},
{
"name": "platformdirs",
"specs": []
},
{
"name": "prometheus_client",
"specs": [
[
"==",
"0.21.1"
]
]
},
{
"name": "prompt-toolkit",
"specs": []
},
{
"name": "psutil",
"specs": []
},
{
"name": "ptyprocess",
"specs": []
},
{
"name": "pure-eval",
"specs": []
},
{
"name": "py_find_1st",
"specs": [
[
"==",
"1.1.7"
]
]
},
{
"name": "pycparser",
"specs": [
[
"==",
"2.22"
]
]
},
{
"name": "Pygments",
"specs": []
},
{
"name": "pynndescent",
"specs": [
[
"==",
"0.5.13"
]
]
},
{
"name": "pyparsing",
"specs": [
[
"==",
"3.2.1"
]
]
},
{
"name": "python-dateutil",
"specs": []
},
{
"name": "python-json-logger",
"specs": [
[
"==",
"3.3.0"
]
]
},
{
"name": "pytz",
"specs": []
},
{
"name": "PyWavelets",
"specs": [
[
"==",
"1.8.0"
]
]
},
{
"name": "PyYAML",
"specs": [
[
"==",
"6.0.2"
]
]
},
{
"name": "pyzmq",
"specs": []
},
{
"name": "referencing",
"specs": [
[
"==",
"0.36.2"
]
]
},
{
"name": "requests",
"specs": [
[
"==",
"2.32.3"
]
]
},
{
"name": "requests-toolbelt",
"specs": [
[
"==",
"1.0.0"
]
]
},
{
"name": "rfc3339-validator",
"specs": [
[
"==",
"0.1.4"
]
]
},
{
"name": "rfc3986-validator",
"specs": [
[
"==",
"0.1.1"
]
]
},
{
"name": "rpds-py",
"specs": [
[
"==",
"0.23.1"
]
]
},
{
"name": "scanpy",
"specs": [
[
"==",
"1.11.0"
]
]
},
{
"name": "scikit-image",
"specs": [
[
"==",
"0.19.3"
]
]
},
{
"name": "scikit-learn",
"specs": [
[
"==",
"1.5.2"
]
]
},
{
"name": "scipy",
"specs": [
[
"==",
"1.15.2"
]
]
},
{
"name": "seaborn",
"specs": [
[
"==",
"0.13.2"
]
]
},
{
"name": "Send2Trash",
"specs": [
[
"==",
"1.8.3"
]
]
},
{
"name": "session-info2",
"specs": [
[
"==",
"0.1.2"
]
]
},
{
"name": "six",
"specs": []
},
{
"name": "sniffio",
"specs": [
[
"==",
"1.3.1"
]
]
},
{
"name": "soupsieve",
"specs": [
[
"==",
"2.6"
]
]
},
{
"name": "stack-data",
"specs": []
},
{
"name": "stardist",
"specs": [
[
"==",
"0.9.1"
]
]
},
{
"name": "statsmodels",
"specs": [
[
"==",
"0.14.4"
]
]
},
{
"name": "terminado",
"specs": [
[
"==",
"0.18.1"
]
]
},
{
"name": "texttable",
"specs": [
[
"==",
"1.7.0"
]
]
},
{
"name": "threadpoolctl",
"specs": [
[
"==",
"3.6.0"
]
]
},
{
"name": "tifffile",
"specs": [
[
"==",
"2025.3.13"
]
]
},
{
"name": "tinycss2",
"specs": [
[
"==",
"1.4.0"
]
]
},
{
"name": "tomli",
"specs": [
[
"==",
"2.2.1"
]
]
},
{
"name": "topact",
"specs": []
},
{
"name": "tornado",
"specs": []
},
{
"name": "tqdm",
"specs": [
[
"==",
"4.67.1"
]
]
},
{
"name": "traitlets",
"specs": []
},
{
"name": "types-python-dateutil",
"specs": [
[
"==",
"2.9.0.20241206"
]
]
},
{
"name": "typing_extensions",
"specs": []
},
{
"name": "tzdata",
"specs": []
},
{
"name": "umap-learn",
"specs": [
[
"==",
"0.5.7"
]
]
},
{
"name": "uri-template",
"specs": [
[
"==",
"1.3.0"
]
]
},
{
"name": "urllib3",
"specs": [
[
"==",
"2.3.0"
]
]
},
{
"name": "wcwidth",
"specs": [
[
"==",
"0.2.13"
]
]
},
{
"name": "webcolors",
"specs": [
[
"==",
"24.11.1"
]
]
},
{
"name": "webencodings",
"specs": [
[
"==",
"0.5.1"
]
]
},
{
"name": "websocket-client",
"specs": [
[
"==",
"1.8.0"
]
]
},
{
"name": "tensorflow",
"specs": [
[
"==",
"2.19.0"
]
]
}
],
"lcname": "spatialcell"
}
